Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR
- PMID: 26238539
- PMCID: PMC4793896
- DOI: 10.1093/bib/bbv056
Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR
Abstract
The high-throughput analysis of microRNAs (miRNAs) circulating within the blood of healthy and diseased individuals is an active area of biomarker research. Whereas quantitative real-time reverse transcription polymerase chain reaction (qPCR)-based methods are widely used, it is yet unresolved how the data should be normalized. Here, we show that a combination of different algorithms results in the identification of candidate reference miRNAs that can be exploited as normalizers, in both discovery and validation phases. Using the methodology considered here, we identify normalizers that are able to reduce nonbiological variation in the data and we present several case studies, to illustrate the relevance in the context of physiological or pathological scenarios. In conclusion, the discovery of stable reference miRNAs from high-throughput studies allows appropriate normalization of focused qPCR assays.
Keywords: Normfinder; circulating miRNA; geNorm; normalization; qPCR; reference genes.
© The Author 2015. Published by Oxford University Press.
Figures
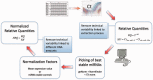

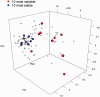
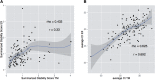
References
-
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. - PubMed
-
- Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 2014;11:809–15. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

