A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis
- PMID: 26240058
- PMCID: PMC5522178
- DOI: 10.1002/art.39287
A longitudinal biomarker for the extent of skin disease in patients with diffuse cutaneous systemic sclerosis
Abstract
Objective: To define a pharmacodynamic biomarker based on gene expression in skin that would provide a biologic measure of the extent of disease in patients with diffuse cutaneous systemic sclerosis (dcSSc) and could be used to monitor skin disease longitudinally.
Methods: Skin biopsy specimens obtained from a cohort of patients with dcSSc (including longitudinal specimens) were analyzed by microarray. Expression of genes correlating with the modified Rodnan skin thickness score (MRSS) were examined for change over time using a NanoString platform, and a generalized estimating equation (GEE) was used to define and validate longitudinally measured pharmacodynamic biomarkers composed of multiple genes.
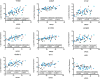
Results: Microarray analysis of genes parsed to include only those correlating with the MRSS revealed prominent clusters of profibrotic/transforming growth factor β-regulated, interferon-regulated/proteasome, macrophage, and vascular marker genes. Using genes changing longitudinally with the MRSS, we defined 2 multigene pharmacodynamic biomarkers. The first was defined mathematically by applying a GEE to longitudinal samples. This modeling method selected cross-sectional THBS1 and longitudinal THBS1 and MS4A4A. The second model was based on a weighted selection of genes, including additional genes that changed statistically significantly over time: CTGF, CD163, CCL2, and WIF1. In an independent validation data set, biomarker levels calculated using both models correlated highly with the MRSS.
Conclusion: Skin gene expression can be used effectively to monitor changes in SSc skin disease over time. We implemented 2 relatively simple models on a NanoString platform permitting highly reproducible assays that can be applied directly to samples from patients or collected as part of clinical trials.
© 2015, American College of Rheumatology.
Figures



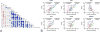

Comment in
-
Connective tissue diseases. Skin gene expression profiles in SSc.Nat Rev Rheumatol. 2015 Oct;11(10):563. doi: 10.1038/nrrheum.2015.115. Epub 2015 Aug 18. Nat Rev Rheumatol. 2015. PMID: 26282083 No abstract available.
References
-
- Gordon JK, Magr C, Udeh U, et al. Nilotinib (Tasigna™) In The Treatment Of Early Diffuse Systemic Sclerosis: A Single Group, Open Label Pilot Clinical Trial – One Year Results. American College of Rheuamtology Meeting Abstracts. 2013:709.
-
- Friendly M. Corrgrams: Exploratory Displays for Correlation Matrices. Tha American Statistician. 2002;56:316–24.
-
- Wright K. Corrgram: Plot a correlogram. R package version 1.6. 2014 http://CRAN.R-project.org/package=corrgram.
-
- McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological Methods. 1996;1:30–46.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

