Reversal of the Drug Binding Pocket Defects of the AcrB Multidrug Efflux Pump Protein of Escherichia coli
- PMID: 26240069
- PMCID: PMC4573724
- DOI: 10.1128/JB.00547-15
Reversal of the Drug Binding Pocket Defects of the AcrB Multidrug Efflux Pump Protein of Escherichia coli
Abstract
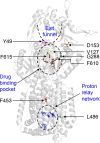
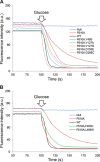
The AcrB protein of Escherichia coli, together with TolC and AcrA, forms a contiguous envelope conduit for the capture and extrusion of diverse antibiotics and cellular metabolites. In this study, we sought to expand our knowledge of AcrB by conducting genetic and functional analyses. We began with an AcrB mutant bearing an F610A substitution in the drug binding pocket and obtained second-site substitutions that overcame the antibiotic hypersusceptibility phenotype conferred by the F610A mutation. Five of the seven unique single amino acid substitutions--Y49S, V127A, V127G, D153E, and G288C--mapped in the periplasmic porter domain of AcrB, with the D153E and G288C mutations mapping near and at the distal drug binding pocket, respectively. The other two substitutions--F453C and L486W--were mapped to transmembrane (TM) helices 5 and 6, respectively. The nitrocefin efflux kinetics data suggested that all periplasmic suppressors significantly restored nitrocefin binding affinity impaired by the F610A mutation. Surprisingly, despite increasing MICs of tested antibiotics and the efflux of N-phenyl-1-naphthylamine, the TM suppressors did not improve the nitrocefin efflux kinetics. These data suggest that the periplasmic substitutions act by influencing drug binding affinities for the distal binding pocket, whereas the TM substitutions may indirectly affect the conformational dynamics of the drug binding domain.
Importance: The AcrB protein and its homologues confer multidrug resistance in many important human bacterial pathogens. A greater understanding of how these efflux pump proteins function will lead to the development of effective inhibitors against them. The research presented in this paper investigates drug binding pocket mutants of AcrB through the isolation and characterization of intragenic suppressor mutations that overcome the drug susceptibility phenotype of mutations affecting the drug binding pocket. The data reveal a remarkable structure-function plasticity of the AcrB protein pertaining to its drug efflux activity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3509-14. doi: 10.1073/pnas.1602472113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976576 Free PMC article.
-
Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB.J Bacteriol. 2008 Dec;190(24):8225-9. doi: 10.1128/JB.00912-08. Epub 2008 Oct 10. J Bacteriol. 2008. PMID: 18849422 Free PMC article.
-
Kinetic analysis of the inhibition of the drug efflux protein AcrB using surface plasmon resonance.Biochim Biophys Acta Biomembr. 2018 Apr;1860(4):878-886. doi: 10.1016/j.bbamem.2017.08.024. Epub 2017 Sep 8. Biochim Biophys Acta Biomembr. 2018. PMID: 28890187
-
The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance.Curr Drug Targets. 2008 Sep;9(9):729-49. doi: 10.2174/138945008785747789. Curr Drug Targets. 2008. PMID: 18781920 Review.
-
Drug transport mechanism of the AcrB efflux pump.Biochim Biophys Acta. 2009 May;1794(5):782-93. doi: 10.1016/j.bbapap.2008.12.015. Epub 2009 Jan 3. Biochim Biophys Acta. 2009. PMID: 19166984 Review.
Cited by
-
Antibiotic Substrate Selectivity of Pseudomonas aeruginosa MexY and MexB Efflux Systems Is Determined by a Goldilocks Affinity.Antimicrob Agents Chemother. 2020 Jul 22;64(8):e00496-20. doi: 10.1128/AAC.00496-20. Print 2020 Jul 22. Antimicrob Agents Chemother. 2020. PMID: 32457110 Free PMC article.
-
Ever-Adapting RND Efflux Pumps in Gram-Negative Multidrug-Resistant Pathogens: A Race against Time.Antibiotics (Basel). 2021 Jun 25;10(7):774. doi: 10.3390/antibiotics10070774. Antibiotics (Basel). 2021. PMID: 34201908 Free PMC article. Review.
-
Using Chemical Probes to Assess the Feasibility of Targeting SecA for Developing Antimicrobial Agents against Gram-Negative Bacteria.ChemMedChem. 2016 Nov 21;11(22):2511-2521. doi: 10.1002/cmdc.201600421. Epub 2016 Oct 18. ChemMedChem. 2016. PMID: 27753464 Free PMC article.
-
Comprehensive Molecular Profiling of AcrAB-TolC Efflux Pump Genes in Salmonella typhi Isolates from Typhoid Infected Patients.Curr Microbiol. 2025 Aug 22;82(10):470. doi: 10.1007/s00284-025-04460-2. Curr Microbiol. 2025. PMID: 40844743
-
Aminoacyl β-naphthylamides as substrates and modulators of AcrB multidrug efflux pump.Proc Natl Acad Sci U S A. 2016 Feb 2;113(5):1405-10. doi: 10.1073/pnas.1525143113. Epub 2016 Jan 19. Proc Natl Acad Sci U S A. 2016. PMID: 26787896 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

