Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi
- PMID: 26240167
- PMCID: PMC4634314
- DOI: 10.1093/glycob/cwv058
Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi
Abstract
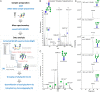
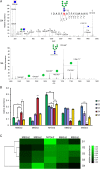
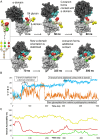
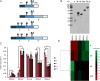
The hallmark of N-linked protein glycosylation is the generation of diverse glycan structures in the secretory pathway. Dynamic, non-template-driven processes of N-glycan remodeling in the endoplasmic reticulum and the Golgi provide the cellular setting for structural diversity. We applied newly developed mass spectrometry-based analytics to quantify site-specific N-glycan remodeling of the model protein Pdi1p expressed in insect cells. Molecular dynamics simulation, mutational analysis, kinetic studies of in vitro processing events and glycan flux analysis supported the defining role of the protein in N-glycan processing.
Keywords: Golgi; Golgi glycosylation; endoplasmic reticulum glycosylation; site-specific glycosylation.
© The Author 2015. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
The Saccharomyces cerevisiae alg12delta mutant reveals a role for the middle-arm alpha1,2Man- and upper-arm alpha1,2Manalpha1,6Man- residues of Glc3Man9GlcNAc2-PP-Dol in regulating glycoprotein glycan processing in the endoplasmic reticulum and Golgi apparatus.Glycobiology. 2002 Nov;12(11):749-62. doi: 10.1093/glycob/cwf082. Glycobiology. 2002. PMID: 12460943
-
Towards understanding the extensive diversity of protein N-glycan structures in eukaryotes.Biol Rev Camb Philos Soc. 2022 Apr;97(2):732-748. doi: 10.1111/brv.12820. Epub 2021 Dec 6. Biol Rev Camb Philos Soc. 2022. PMID: 34873817 Free PMC article. Review.
-
Influence of protein/glycan interaction on site-specific glycan heterogeneity.FASEB J. 2017 Oct;31(10):4623-4635. doi: 10.1096/fj.201700403R. Epub 2017 Jul 5. FASEB J. 2017. PMID: 28679530
-
Plant glyco-biotechnology.Semin Cell Dev Biol. 2018 Aug;80:133-141. doi: 10.1016/j.semcdb.2017.07.005. Epub 2017 Jul 5. Semin Cell Dev Biol. 2018. PMID: 28688929 Review.
-
Biosynthesis of the side chain of yeast glycosylphosphatidylinositol anchors is operated by novel mannosyltransferases located in the endoplasmic reticulum and the Golgi apparatus.J Biol Chem. 1995 Aug 25;270(34):19709-15. doi: 10.1074/jbc.270.34.19709. J Biol Chem. 1995. PMID: 7649981
Cited by
-
The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.J Am Chem Soc. 2016 Jun 22;138(24):7636-48. doi: 10.1021/jacs.6b02879. Epub 2016 Jun 14. J Am Chem Soc. 2016. PMID: 27249581 Free PMC article.
-
The N-Glycosylation Processing Potential of the Mammalian Golgi Apparatus.Front Cell Dev Biol. 2019 Aug 13;7:157. doi: 10.3389/fcell.2019.00157. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31457009 Free PMC article.
-
An overview on glycation: molecular mechanisms, impact on proteins, pathogenesis, and inhibition.Biophys Rev. 2024 Apr 12;16(2):189-218. doi: 10.1007/s12551-024-01188-4. eCollection 2024 Apr. Biophys Rev. 2024. PMID: 38737201 Free PMC article. Review.
-
Substrate recognition and catalysis by GH47 α-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.Proc Natl Acad Sci U S A. 2016 Dec 6;113(49):E7890-E7899. doi: 10.1073/pnas.1611213113. Epub 2016 Nov 17. Proc Natl Acad Sci U S A. 2016. PMID: 27856750 Free PMC article.
-
Comparison of pregnant mare serum gonadotropin products with surprising differences in protein content.Sci Rep. 2025 Feb 25;15(1):6824. doi: 10.1038/s41598-025-90833-3. Sci Rep. 2025. PMID: 40000800 Free PMC article.
References
-
- Aebi M, Bernasconi R, Clerc S, Molinari M. 2010. N-Glycan structures: Recognition and processing in the ER. Trends Biochem Sci. 35:74–82. - PubMed
-
- Antoniewicz MR. 2013. Dynamic metabolic flux analysis – Tools for probing transient states of metabolic networks. Curr Opin Biotechnol. 24:973–978. - PubMed
-
- Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H et al. . 2014. AMBER 14. San Francisco: University of California.
-
- Darden T, York D, Pedersen L. 1993. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J Chem Phys. 98:10089.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

