Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily
- PMID: 26240321
- PMCID: PMC4577193
- DOI: 10.1073/pnas.1500964112
Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily
Abstract
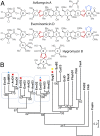
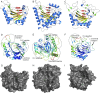
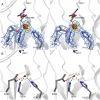
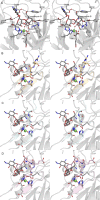
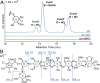
Orthosomycins are oligosaccharide antibiotics that include avilamycin, everninomicin, and hygromycin B and are hallmarked by a rigidifying interglycosidic spirocyclic ortho-δ-lactone (orthoester) linkage between at least one pair of carbohydrates. A subset of orthosomycins additionally contain a carbohydrate capped by a methylenedioxy bridge. The orthoester linkage is necessary for antibiotic activity but rarely observed in natural products. Orthoester linkage and methylenedioxy bridge biosynthesis require similar oxidative cyclizations adjacent to a sugar ring. We have identified a conserved group of nonheme iron, α-ketoglutarate-dependent oxygenases likely responsible for this chemistry. High-resolution crystal structures of the EvdO1 and EvdO2 oxygenases of everninomicin biosynthesis, the AviO1 oxygenase of avilamycin biosynthesis, and HygX of hygromycin B biosynthesis show how these enzymes accommodate large substrates, a challenge that requires a variation in metal coordination in HygX. Excitingly, the ternary complex of HygX with cosubstrate α-ketoglutarate and putative product hygromycin B identified an orientation of one glycosidic linkage of hygromycin B consistent with metal-catalyzed hydrogen atom abstraction from substrate. These structural results are complemented by gene disruption of the oxygenases evdO1 and evdMO1 from the everninomicin biosynthetic cluster, which demonstrate that functional oxygenase activity is critical for antibiotic production. Our data therefore support a role for these enzymes in the production of key features of the orthosomycin antibiotics.
Keywords: antibiotic biosynthesis; crystal structure; nonheme iron α-ketoglutarate–dependent oxygenases; oxidative cyclization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




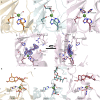




Comment in
-
Assembly of the unusual oxacycles in the orthosomycin antibiotics.Proc Natl Acad Sci U S A. 2015 Sep 29;112(39):11989-90. doi: 10.1073/pnas.1514689112. Epub 2015 Sep 16. Proc Natl Acad Sci U S A. 2015. PMID: 26378126 Free PMC article. No abstract available.
References
-
- Jones RN, Low DE, Pfaller MA. Epidemiologic trends in nosocomial and community-acquired infections due to antibiotic-resistant gram-positive bacteria: The role of streptogramins and other newer compounds. Diagn Microbiol Infect Dis. 1999;33(2):101–112. - PubMed
-
- Waitz JA, Horan AC. 1988. Micromonospora carbonacea var africana. US Patent 4,735,903.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

