Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations
- PMID: 26240355
- PMCID: PMC4568650
- DOI: 10.1073/pnas.1506184112
Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations
Abstract
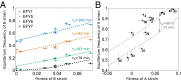
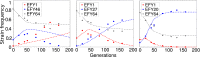
Identifying the mechanisms that create and maintain biodiversity is a central challenge in biology. Stable diversification of microbial populations often requires the evolution of differences in resource utilization. Alternatively, coexistence can be maintained by specialization to exploit spatial heterogeneity in the environment. Here, we report spontaneous diversification maintained by a related but distinct mechanism: crowding avoidance. During experimental evolution of laboratory Saccharomyces cerevisiae populations, we observed the repeated appearance of "adherent" (A) lineages able to grow as a dispersed film, in contrast to their crowded "bottom-dweller" (B) ancestors. These two types stably coexist because dispersal reduces interference competition for nutrients among kin, at the cost of a slower maximum growth rate. This tradeoff causes the frequencies of the two types to oscillate around equilibrium over the course of repeated cycles of growth, crowding, and dispersal. However, further coevolution of the A and B types can perturb and eventually destroy their coexistence over longer time scales. We introduce a simple mathematical model of this "semistable" coexistence, which explains the interplay between ecological and evolutionary dynamics. Because crowded growth generally limits nutrient access in biofilms, the mechanism we report here may be broadly important in maintaining diversity in these natural environments.
Keywords: coexistence; crowding; experimental evolution; fungal adherence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
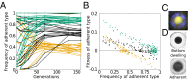
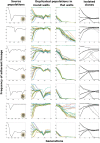
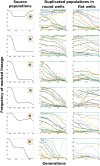

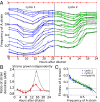
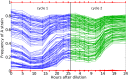
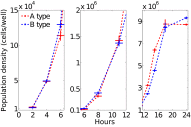

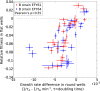


Comment in
-
Evolution of coexistence in a crowded microplate well.Proc Natl Acad Sci U S A. 2015 Sep 8;112(36):11148-9. doi: 10.1073/pnas.1514399112. Epub 2015 Aug 26. Proc Natl Acad Sci U S A. 2015. PMID: 26311847 Free PMC article. No abstract available.
References
-
- Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58(2):245–260. - PubMed
-
- Craig MacLean R, Dickson A, Bell G. Resource competition and adaptive radiation in a microbial microcosm. Ecol Lett. 2005;8(1):38–46.
-
- Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett. 2009;12(1):34–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

