Application of the consolidated species concept to Cercospora spp. from Iran
- PMID: 26240446
- PMCID: PMC4510272
- DOI: 10.3767/003158515X685698
Application of the consolidated species concept to Cercospora spp. from Iran
Abstract
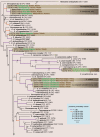
The genus Cercospora includes many important plant pathogenic fungi associated with leaf spot diseases on a wide range of hosts. The mainland of Iran covers various climatic regions with a great biodiversity of vascular plants, and a correspondingly high diversity of cercosporoid fungi. However, most of the cercosporoid species found to date have been identified on the basis of morphological characteristics and there are no cultures that support these identifications. In this study the Consolidated Species Concept was applied to differentiate Cercospora species collected from Iran. A total of 161 Cercospora isolates recovered from 74 host species in northern Iran were studied by molecular phylogenetic analysis. Our results revealed a rich diversity of Cercospora species in northern Iran. Twenty species were identified based on sequence data of five genomic loci (ITS, TEF1-α, actin, calmodulin and histone H3), host, cultural and morphological data. Six novel species, viz. C. convolvulicola, C. conyzae-canadensis, C. cylindracea, C. iranica, C. pseudochenopodii and C. sorghicola, are introduced. The most common taxon was Cercospora cf. flagellaris, which remains an unresolved species complex with a wide host range. New hosts were recorded for previously known Cercospora species, including C. apii, C. armoraciae, C. beticola, C. cf. richardiicola, C. rumicis, Cercospora sp. G and C. zebrina.
Keywords: Cercospora apii complex; Mycosphaerella; biodiversity; cercosporoid; host specificity; leaf spot; multilocus sequence typing (MLST); taxonomy.
Figures














References
-
- Agrios GN. 2005. Plant pathology, fifth edition Academic Press, New York.
-
- Aptroot A. 2006. Mycosphaerella and its anamorphs: 2. Conspectus of Mycosphaerella. CBS Biodiversity Series 5: 1–231.
-
- Arzanlou M, Abeln EC, Kema GH, et al. 2007. Molecular diagnostics for the Sigatoka disease complex of banana. Phytopathology 97: 1112–1118. - PubMed
-
- Bakhshi M, Arzanlou M, Babai-Ahari A. 2011. Uneven distribution of mating type alleles in Iranian populations of Cercospora beticola, the causal agent of Cercospora leaf spot disease of sugar beet. Phytopathologia Mediterranea 50: 101–109.
LinkOut - more resources
Full Text Sources
Miscellaneous
