Structural insights into the regulation of aromatic amino acid hydroxylation
- PMID: 26241318
- PMCID: PMC4688188
- DOI: 10.1016/j.sbi.2015.07.004
Structural insights into the regulation of aromatic amino acid hydroxylation
Abstract
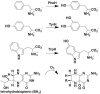
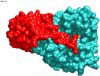
The aromatic amino acid hydroxylases phenylalanine hydroxylase, tyrosine hydroxylase, and tryptophan hydroxylase are homotetramers, with each subunit containing a homologous catalytic domain and a divergent regulatory domain. The solution structure of the regulatory domain of tyrosine hydroxylase establishes that it contains a core ACT domain similar to that in phenylalanine hydroxylase. The isolated regulatory domain of tyrosine hydroxylase forms a stable dimer, while that of phenylalanine hydroxylase undergoes a monomer-dimer equilibrium, with phenylalanine stabilizing the dimer. These solution properties are consistent with the regulatory mechanisms of the two enzymes, in that phenylalanine hydroxylase is activated by phenylalanine binding to an allosteric site, while tyrosine hydroxylase is regulated by binding of catecholamines in the active site.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures






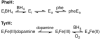
References
-
- Fitzpatrick PF. Tetrahydropterin-dependent amino acid hydroxylases. Ann. Rev. Biochem. 1999;68:355–381. - PubMed
-
- Lewis DA, Melchitzky DS, Haycock JW. Four isoforms of tyrosine hydroxylase are expressed in human brain. Neuroscience. 1993;54:477–492. - PubMed
-
- Le Bourdellès B, Horellou P, Le Caer J-P, Denèfle P, Latta M, Haavik J, Guibert B, Mayaux J-F, Mallet J. Phosphorylation of human recombinant tyrosine hydroxylase isoforms 1 and 2: An additional phosphorylated residue in isoform 2, generated through alternative splicing. J. Biol. Chem. 1991;266:17124–17130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

