The methylaspartate cycle in haloarchaea and its possible role in carbon metabolism
- PMID: 26241502
- PMCID: PMC4817688
- DOI: 10.1038/ismej.2015.132
The methylaspartate cycle in haloarchaea and its possible role in carbon metabolism
Abstract
Haloarchaea (class Halobacteria) live in extremely halophilic conditions and evolved many unique metabolic features, which help them to adapt to their environment. The methylaspartate cycle, an anaplerotic acetate assimilation pathway recently proposed for Haloarcula marismortui, is one of these special adaptations. In this cycle, acetyl-CoA is oxidized to glyoxylate via methylaspartate as a characteristic intermediate. The following glyoxylate condensation with another molecule of acetyl-CoA yields malate, a starting substrate for anabolism. The proposal of the functioning of the cycle was based mainly on in vitro data, leaving several open questions concerning the enzymology involved and the occurrence of the cycle in halophilic archaea. Using gene deletion mutants of H. hispanica, enzyme assays and metabolite analysis, we now close these gaps by unambiguous identification of the genes encoding all characteristic enzymes of the cycle. Based on these results, we were able to perform a solid study of the distribution of the methylaspartate cycle and the alternative acetate assimilation strategy, the glyoxylate cycle, among haloarchaea. We found that both of these cycles are evenly distributed in haloarchaea. Interestingly, 83% of the species using the methylaspartate cycle possess also the genes for polyhydroxyalkanoate biosynthesis, whereas only 34% of the species with the glyoxylate cycle are capable to synthesize this storage compound. This finding suggests that the methylaspartate cycle is shaped for polyhydroxyalkanoate utilization during carbon starvation, whereas the glyoxylate cycle is probably adapted for growth on substrates metabolized via acetyl-CoA.
Figures
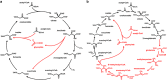
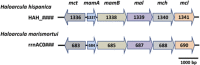
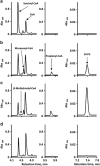
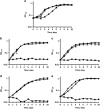
 ), pyruvate (
), pyruvate ( ) or acetate and pyruvate (
) or acetate and pyruvate ( ). The experiment was done in triplicate, the error bars represent the standard deviations. Note that the mutants did not grow on the medium with acetate alone even after 21 days of incubation. The Δmct mutant grows slowly on the medium with pyruvate compared with the wild-type strain. The reason for that is unknown.
). The experiment was done in triplicate, the error bars represent the standard deviations. Note that the mutants did not grow on the medium with acetate alone even after 21 days of incubation. The Δmct mutant grows slowly on the medium with pyruvate compared with the wild-type strain. The reason for that is unknown.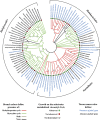
Similar articles
-
Malate Synthase and β-Methylmalyl Coenzyme A Lyase Reactions in the Methylaspartate Cycle in Haloarcula hispanica.J Bacteriol. 2017 Jan 30;199(4):e00657-16. doi: 10.1128/JB.00657-16. Print 2017 Feb 15. J Bacteriol. 2017. PMID: 27920298 Free PMC article.
-
A methylaspartate cycle in haloarchaea.Science. 2011 Jan 21;331(6015):334-7. doi: 10.1126/science.1196544. Science. 2011. PMID: 21252347
-
Succinyl-CoA:Mesaconate CoA-Transferase and Mesaconyl-CoA Hydratase, Enzymes of the Methylaspartate Cycle in Haloarcula hispanica.Front Microbiol. 2017 Sep 6;8:1683. doi: 10.3389/fmicb.2017.01683. eCollection 2017. Front Microbiol. 2017. PMID: 28932214 Free PMC article.
-
Revisiting the glyoxylate cycle: alternate pathways for microbial acetate assimilation.Mol Microbiol. 2006 Jul;61(2):274-6. doi: 10.1111/j.1365-2958.2006.05247.x. Mol Microbiol. 2006. PMID: 16856935 Review.
-
Unfamiliar metabolic links in the central carbon metabolism.J Biotechnol. 2014 Dec 20;192 Pt B:314-22. doi: 10.1016/j.jbiotec.2014.02.015. Epub 2014 Feb 24. J Biotechnol. 2014. PMID: 24576434 Review.
Cited by
-
With a pinch of salt: metagenomic insights into Namib Desert salt pan microbial mats and halites reveal functionally adapted and competitive communities.Appl Environ Microbiol. 2023 Dec 21;89(12):e0062923. doi: 10.1128/aem.00629-23. Epub 2023 Nov 16. Appl Environ Microbiol. 2023. PMID: 37971255 Free PMC article.
-
Pseudomonadal itaconate degradation gene cluster encodes enzymes for methylsuccinate utilization.Commun Biol. 2025 Jul 24;8(1):1099. doi: 10.1038/s42003-025-08538-2. Commun Biol. 2025. PMID: 40707647 Free PMC article.
-
Acetate Metabolism in Archaea: Characterization of an Acetate Transporter and of Enzymes Involved in Acetate Activation and Gluconeogenesis in Haloferax volcanii.Front Microbiol. 2020 Dec 4;11:604926. doi: 10.3389/fmicb.2020.604926. eCollection 2020. Front Microbiol. 2020. PMID: 33343547 Free PMC article.
-
TCA Cycle Replenishing Pathways in Photosynthetic Purple Non-Sulfur Bacteria Growing with Acetate.Life (Basel). 2021 Jul 19;11(7):711. doi: 10.3390/life11070711. Life (Basel). 2021. PMID: 34357087 Free PMC article. Review.
-
Propionate metabolism in Desulfurella acetivorans.Front Microbiol. 2025 Feb 12;16:1545849. doi: 10.3389/fmicb.2025.1545849. eCollection 2025. Front Microbiol. 2025. PMID: 40012784 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Bergmeyer HU. (1975). Neue Werte für die molaren Extinktions-Koeffizienten von NADH und NADPH zum Gebrauch im Routine-Laboratorium. Z Klin Chem Klin Biochem 13: 507–508. - PubMed
-
- Bradford MM. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

