Competitive strategies differentiate closely related species of marine actinobacteria
- PMID: 26241505
- PMCID: PMC4737938
- DOI: 10.1038/ismej.2015.128
Competitive strategies differentiate closely related species of marine actinobacteria
Abstract
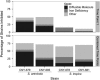
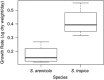
Although competition, niche partitioning, and spatial isolation have been used to describe the ecology and evolution of macro-organisms, it is less clear to what extent these principles account for the extraordinary levels of bacterial diversity observed in nature. Ecological interactions among bacteria are particularly challenging to address due to methodological limitations and uncertainties over how to recognize fundamental units of diversity and link them to the functional traits and evolutionary processes that led to their divergence. Here we show that two closely related marine actinomycete species can be differentiated based on competitive strategies. Using a direct challenge assay to investigate inhibitory interactions with members of the bacterial community, we observed a temporal difference in the onset of inhibition. The majority of inhibitory activity exhibited by Salinispora arenicola occurred early in its growth cycle and was linked to antibiotic production. In contrast, most inhibition by Salinispora tropica occurred later in the growth cycle and was more commonly linked to nutrient depletion or other sources. Comparative genomics support these differences, with S. arenicola containing nearly twice the number of secondary metabolite biosynthetic gene clusters as S. tropica, indicating a greater potential for secondary metabolite production. In contrast, S. tropica is enriched in gene clusters associated with the acquisition of growth-limiting nutrients such as iron. Coupled with differences in growth rates, the results reveal that S. arenicola uses interference competition at the expense of growth, whereas S. tropica preferentially employs a strategy of exploitation competition. The results support the ecological divergence of two co-occurring and closely related species of marine bacteria by providing evidence they have evolved fundamentally different strategies to compete in marine sediments.
Figures




References
-
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430: 551–554. - PubMed
-
- Ahmed L, Jensen PR, Freel KC, Brown R, Jones AL, Kim BY et al. (2013). Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie Van Leeuwenhoek 103: 1069–1078. - PubMed
-
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
-
- Bellenger JP, Wichard T, Kustka AB, Kraepiel AML. (2008). Uptake of molybdenum and vanadium by a nitrogen-fixing soil bacterium using siderophores. Nat Geosci 1: 243–246.
-
- Berdy J. (2005). Bioactive microbial metabolites. A personal view. J Antibiot 58: 1–26. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

