Genetic determinants of FOXM1 overexpression in epithelial ovarian cancer and functional contribution to cell cycle progression
- PMID: 26243836
- PMCID: PMC4695012
- DOI: 10.18632/oncotarget.4546
Genetic determinants of FOXM1 overexpression in epithelial ovarian cancer and functional contribution to cell cycle progression
Abstract
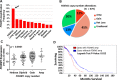
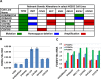
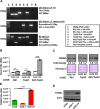
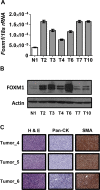
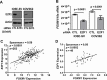
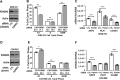
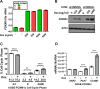
The FOXM1 transcription factor network is frequently activated in high-grade serous ovarian cancer (HGSOC), the most common and lethal subtype of epithelial ovarian cancer (EOC). We used primary human EOC tissues, HGSOC cell lines, mouse and human ovarian surface epithelial (OSE) cells, and a murine transgenic ovarian cancer model to investigate genetic determinants of FOXM1 overexpression in EOC, and to begin to define its functional contribution to disease pathology. The Cancer Genome Atlas (TCGA) data indicated that the FOXM1 locus is amplified in ~12% of HGSOC, greater than any other tumor type examined, and that FOXM1 amplification correlates with increased expression and poor survival. In an independent set of primary EOC tissues, FOXM1 expression correlated with advanced stage and grade. Of the three known FOXM1 isoforms, FOXM1c showed highest expression in EOC. In murine OSE cells, combined knockout of Rb1 and Trp53 synergistically induced FOXM1. Consistently, human OSE cells immortalized with SV40 Large T antigen (IOSE-SV) had significantly higher FOXM1 expression than OSE immortalized with hTERT (IOSE-T). FOXM1 was overexpressed in murine ovarian tumors driven by combined Rb1/Trp53 disruption. FOXM1 induction in IOSE-SV cells was partially dependent on E2F1, and FOXM1 expression correlated with E2F1 expression in human EOC tissues. Finally, FOXM1 functionally contributed to cell cycle progression and relevant target gene expression in human OSE and HGSOC cell models. In summary, gene amplification, p53 and Rb disruption, and E2F1 activation drive FOXM1 expression in EOC, and FOXM1 promotes cell cycle progression in EOC cell models.
Keywords: E2F1; FOXM1; Rb; epithelial ovarian cancer; p53.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures

References
-
- Cannistra S.A. Cancer of the ovary. The New England journal of medicine. 2004;351:2519–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

