Antibiotic susceptibility and heterogeneity in technological traits of lactobacilli isolated from Algerian goat's milk
- PMID: 26243893
- PMCID: PMC4519446
- DOI: 10.1007/s13197-014-1556-7
Antibiotic susceptibility and heterogeneity in technological traits of lactobacilli isolated from Algerian goat's milk
Abstract
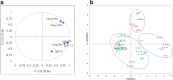
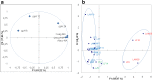
The objective of this study was to identify and study the heterogeneity of technological traits of lactobacilli from goat's milk of Algeria and to evaluate in vitro their safety aspect. Using API50 CHL system and 16S rDNA sequencing, 51 % of strains were assigned as Lactobacillus plantarum, 34 % as L. pentosus, 7 % as L. rhamnosus and 8 % as L. fermentum. A large variability was noted for the acidifying capacity in skim milk after 6, 12 and 24 h of incubation. All strains expressed aminopeptidase activity against alanine-ρ-NA and leucine-ρ-NA at different levels. All strains were resistant to vancomycin and most of strains showed more susceptibility to β-lactam antibiotic. High susceptibility toward the inhibitors of protein synthesis was also observed. Minimum inhibitory concentrations data obtained revealed that isolates were susceptible to penicillin and chloramphenicol, and resistant to gentamicin and vancomycin. Minimum inhibitory concentrations distribution of other antibiotics showed variability. The analysis of graphical representation of principal component analysis of technological properties of L. plantarum and L. pentosus strains showed diversity among the isolates. Finally, eight L. plantarum (LAM1, LAM3, LAM21, LAM25, LAM35, LF15, LAM34, and LAM35), four L. pentosus (LAM38, LAM39, LF9 and LF16) and two L. rhamnosus (LF3 and LF10) strains, could be good candidates as adjunct culture in dairy product in Algeria.
Keywords: Algeria; Antibiotic susceptibility; Goat’s milk; Heterogeneity; Lactobacilli; Technological properties.
Figures

References
-
- Ayad EHE, Nashat S, El-Sadek N, Metwaly H, El-Soda M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004;21:715–725. doi: 10.1016/j.fm.2004.02.009. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
