Effective Population Size and Signatures of Selection Using Bovine 50K SNP Chips in Korean Native Cattle (Hanwoo)
- PMID: 26244003
- PMCID: PMC4498654
- DOI: 10.4137/EBO.S24359
Effective Population Size and Signatures of Selection Using Bovine 50K SNP Chips in Korean Native Cattle (Hanwoo)
Abstract
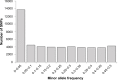
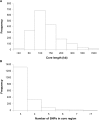
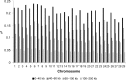
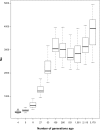
Inferring the effective population size and the pattern of selection signatures is of interest both from an evolutionary perspective and to improve models for mapping of quantitative trait genes. We used DNA samples of 61 sires and 486 progeny of the Hanwoo, genotyped by the Illumina Bovine SNP50 BeadChip, to analyze the genetic structure. Our study showed a persistent decline in effective population size throughout the period considered, but suggested a marked decline at one distinctive time point (100th generation) and two sharp decline intervals (50th-25th generation and 25th-10th generation). This pattern can be explained by Hanwoo formation and the modern breeding program. Our results revealed 95 regions exhibiting the footprint of recent positive selection at a threshold level of 0.01. We found an overlap of the 11 core regions presenting top P-values and those that had previously been identified as harboring quantitative trait loci from other breeds. The information generated from this study can be used to better understand the mechanism of selection in Hanwoo breeding, and provide important implications for the design and application of association studies in the Hanwoo population.
Keywords: cattle; effective population size; linkage disequilibrium; selection signature.
Figures

Similar articles
-
Demographic Trends in Korean Native Cattle Explained Using Bovine SNP50 Beadchip.Genomics Inform. 2016 Dec;14(4):230-233. doi: 10.5808/GI.2016.14.4.230. Epub 2016 Dec 31. Genomics Inform. 2016. PMID: 28154516 Free PMC article.
-
Long-term artificial selection of Hanwoo (Korean) cattle left genetic signatures for the breeding traits and has altered the genomic structure.Sci Rep. 2022 Apr 19;12(1):6438. doi: 10.1038/s41598-022-09425-0. Sci Rep. 2022. PMID: 35440706 Free PMC article.
-
Multiple Linkage Disequilibrium Mapping Methods to Validate Additive Quantitative Trait Loci in Korean Native Cattle (Hanwoo).Asian-Australas J Anim Sci. 2015 Jul;28(7):926-35. doi: 10.5713/ajas.15.0077. Asian-Australas J Anim Sci. 2015. PMID: 26104396 Free PMC article.
-
Genome-wide association study and prediction of genomic breeding values for fatty-acid composition in Korean Hanwoo cattle using a high-density single-nucleotide polymorphism array.J Anim Sci. 2018 Sep 29;96(10):4063-4075. doi: 10.1093/jas/sky280. J Anim Sci. 2018. PMID: 30265318 Free PMC article.
-
Hanwoo cattle: origin, domestication, breeding strategies and genomic selection.J Anim Sci Technol. 2014 May 15;56:2. doi: 10.1186/2055-0391-56-2. eCollection 2014. J Anim Sci Technol. 2014. PMID: 26290691 Free PMC article. Review.
Cited by
-
Genomic prediction for growth using a low-density SNP panel in dromedary camels.Sci Rep. 2021 Apr 7;11(1):7675. doi: 10.1038/s41598-021-87296-7. Sci Rep. 2021. PMID: 33828208 Free PMC article.
-
Demographic Trends in Korean Native Cattle Explained Using Bovine SNP50 Beadchip.Genomics Inform. 2016 Dec;14(4):230-233. doi: 10.5808/GI.2016.14.4.230. Epub 2016 Dec 31. Genomics Inform. 2016. PMID: 28154516 Free PMC article.
-
Genetic characteristics of Korean Jeju Black cattle with high density single nucleotide polymorphisms.Anim Biosci. 2021 May;34(5):789-800. doi: 10.5713/ajas.19.0888. Epub 2020 Aug 21. Anim Biosci. 2021. PMID: 32882779 Free PMC article.
-
Predictive performance of genomic selection methods for carcass traits in Hanwoo beef cattle: impacts of the genetic architecture.Genet Sel Evol. 2017 Jan 4;49(1):1. doi: 10.1186/s12711-016-0283-0. Genet Sel Evol. 2017. PMID: 28093066 Free PMC article.
-
Comparative assessment of the effective population size and linkage disequilibrium of Karan Fries cattle revealed viable population dynamics.Anim Biosci. 2024 May;37(5):795-806. doi: 10.5713/ab.23.0263. Epub 2023 Nov 2. Anim Biosci. 2024. PMID: 37946419 Free PMC article.
References
-
- Habier D. More than a third of the WCGALP presentations on genomic selection. J Anim Breed Genet. 2010;127(5):336–7. - PubMed
-
- Goddard ME, Hayes BJ. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet. 2009;10(6):381–91. - PubMed
-
- Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. - PubMed
-
- Park L. Effective population size of current human population. Genet Res (Camb) 2011;93(2):105–14. - PubMed
LinkOut - more resources
Full Text Sources

