Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis
- PMID: 26244061
- PMCID: PMC4524428
- DOI: 10.1186/s13148-015-0104-2
Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis
Abstract
Background: Interest in the potential of DNA methylation in peripheral blood as a biomarker of cancer risk is increasing. We aimed to assess whether epigenome-wide DNA methylation measured in peripheral blood samples obtained before onset of the disease is associated with increased risk of breast cancer. We report on three independent prospective nested case-control studies from the European Prospective Investigation into Cancer and Nutrition (EPIC-Italy; n = 162 matched case-control pairs), the Norwegian Women and Cancer study (NOWAC; n = 168 matched pairs), and the Breakthrough Generations Study (BGS; n = 548 matched pairs). We used the Illumina 450k array to measure methylation in the EPIC and NOWAC cohorts. Whole-genome bisulphite sequencing (WGBS) was performed on the BGS cohort using pooled DNA samples, combined to reach 50× coverage across ~16 million CpG sites in the genome including 450k array CpG sites. Mean β values over all probes were calculated as a measurement for epigenome-wide methylation.
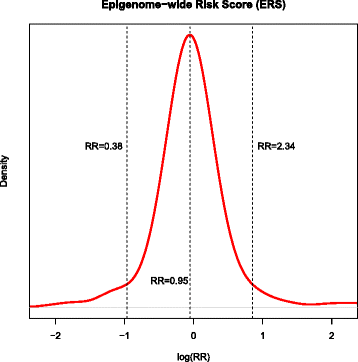
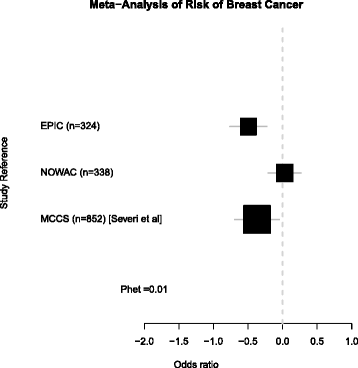
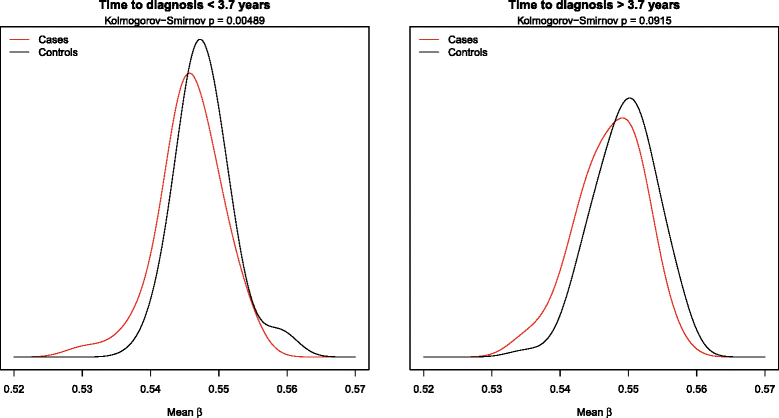
Results: In EPIC, we found that high epigenome-wide methylation was associated with lower risk of breast cancer (odds ratio (OR) per 1 SD = 0.61, 95 % confidence interval (CI) 0.47-0.80; -0.2 % average difference in epigenome-wide methylation for cases and controls). Specifically, this was observed in gene bodies (OR = 0.51, 95 % CI 0.38-0.69) but not in gene promoters (OR = 0.92, 95 % CI 0.64-1.32). The association was not replicated in NOWAC (OR = 1.03 95 % CI 0.81-1.30). The reasons for heterogeneity across studies are unclear. However, data from the BGS cohort was consistent with epigenome-wide hypomethylation in breast cancer cases across the overlapping 450k probe sites (difference in average epigenome-wide methylation in case and control DNA pools = -0.2 %).
Conclusions: We conclude that epigenome-wide hypomethylation of DNA from pre-diagnostic blood samples may be predictive of breast cancer risk and may thus be useful as a clinical biomarker.
Keywords: Biomarker; Breast cancer; EWAS; Methylation; Peripheral blood; Risk.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

