Novel Histone Deacetylase Class IIa Selective Substrate Radiotracers for PET Imaging of Epigenetic Regulation in the Brain
- PMID: 26244761
- PMCID: PMC4526562
- DOI: 10.1371/journal.pone.0133512
Novel Histone Deacetylase Class IIa Selective Substrate Radiotracers for PET Imaging of Epigenetic Regulation in the Brain
Abstract
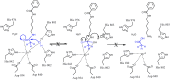
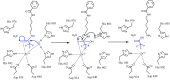
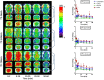
Histone deacetylases (HDAC's) became increasingly important targets for therapy of various diseases, resulting in a pressing need to develop HDAC class- and isoform-selective inhibitors. Class IIa deacetylases possess only minimal deacetylase activity against acetylated histones, but have several other client proteins as substrates through which they participate in epigenetic regulation. Herein, we report the radiosyntheses of the second generation of HDAC class IIa-specific radiotracers: 6-(di-fluoroacetamido)-1-hexanoicanilide (DFAHA) and 6-(tri-fluoroacetamido)-1-hexanoicanilide ([18F]-TFAHA). The selectivity of these radiotracer substrates to HDAC class IIa enzymes was assessed in vitro, in a panel of recombinant HDACs, and in vivo using PET/CT imaging in rats. [18F]TFAHA showed significantly higher selectivity for HDAC class IIa enzymes, as compared to [18F]DFAHA and previously reported [18F]FAHA. PET imaging with [18F]TFAHA can be used to visualize and quantify spatial distribution and magnitude of HDAC class IIa expression-activity in different organs and tissues in vivo. Furthermore, PET imaging with [18F]TFAHA may advance the understanding of HDACs class IIa mediated epigenetic regulation of normal and pathophysiological processes, and facilitate the development of novel HDAC class IIa-specific inhibitors for therapy of different diseases.
Conflict of interest statement
Figures








References
-
- Park JH, Jung Y, Kim TY, Kim SG, Jong HS, et al. (2004) Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation. Clin Cancer Res 10: 5271–5281. - PubMed
-
- Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D (2001) Functional significance of histone deacetylase diversity. Curr Opin Genet Dev 11: 162–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

