Hepatitis C Virus Envelope Glycoprotein E1 Forms Trimers at the Surface of the Virion
- PMID: 26246575
- PMCID: PMC4580159
- DOI: 10.1128/JVI.00991-15
Hepatitis C Virus Envelope Glycoprotein E1 Forms Trimers at the Surface of the Virion
Abstract
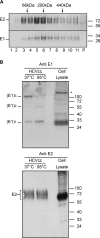
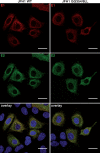
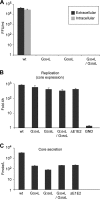
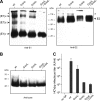
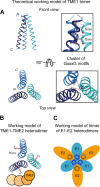
In hepatitis C virus (HCV)-infected cells, the envelope glycoproteins E1 and E2 assemble as a heterodimer. To investigate potential changes in the oligomerization of virion-associated envelope proteins, we performed SDS-PAGE under reducing conditions but without thermal denaturation. This revealed the presence of SDS-resistant trimers of E1 in the context of cell-cultured HCV (HCVcc) as well as in the context of HCV pseudoparticles (HCVpp). The formation of E1 trimers was found to depend on the coexpression of E2. To further understand the origin of E1 trimer formation, we coexpressed in bacteria the transmembrane (TM) domains of E1 (TME1) and E2 (TME2) fused to reporter proteins and analyzed the fusion proteins by SDS-PAGE and Western blotting. As expected for strongly interacting TM domains, TME1-TME2 heterodimers resistant to SDS were observed. These analyses also revealed homodimers and homotrimers of TME1, indicating that such complexes are stable species. The N-terminal segment of TME1 exhibits a highly conserved GxxxG sequence, a motif that is well documented to be involved in intramembrane protein-protein interactions. Single or double mutations of the glycine residues (Gly354 and Gly358) in this motif markedly decreased or abrogated the formation of TME1 homotrimers in bacteria, as well as homotrimers of E1 in both HCVpp and HCVcc systems. A concomitant loss of infectivity was observed, indicating that the trimeric form of E1 is essential for virus infectivity. Taken together, these results indicate that E1E2 heterodimers form trimers on HCV particles, and they support the hypothesis that E1 could be a fusion protein.
Importance: HCV glycoproteins E1 and E2 play an essential role in virus entry into liver cells as well as in virion morphogenesis. In infected cells, these two proteins form a complex in which E2 interacts with cellular receptors, whereas the function of E1 remains poorly understood. However, recent structural data suggest that E1 could be the protein responsible for the process of fusion between viral and cellular membranes. Here we investigated the oligomeric state of HCV envelope glycoproteins. We demonstrate that E1 forms functional trimers after virion assembly and that in addition to the requirement for E2, a determinant for this oligomerization is present in a conserved GxxxG motif located within the E1 transmembrane domain. Taken together, these results indicate that a rearrangement of E1E2 heterodimer complexes likely occurs during the assembly of HCV particles to yield a trimeric form of the E1E2 heterodimer. Gaining structural information on this trimer will be helpful for the design of an anti-HCV vaccine.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry.J Virol. 2007 Mar;81(5):2372-81. doi: 10.1128/JVI.02198-06. Epub 2006 Dec 13. J Virol. 2007. PMID: 17166909 Free PMC article.
-
Identification of new functional regions in hepatitis C virus envelope glycoprotein E2.J Virol. 2011 Feb;85(4):1777-92. doi: 10.1128/JVI.02170-10. Epub 2010 Dec 8. J Virol. 2011. PMID: 21147916 Free PMC article.
-
Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry.Hepatology. 2014 Mar;59(3):776-88. doi: 10.1002/hep.26733. Epub 2014 Jan 28. Hepatology. 2014. PMID: 24038151
-
HCV glycoprotein structures: what to expect from the unexpected.Curr Opin Virol. 2015 Jun;12:53-8. doi: 10.1016/j.coviro.2015.02.004. Epub 2015 Mar 16. Curr Opin Virol. 2015. PMID: 25790756 Free PMC article. Review.
-
Assembly of a functional HCV glycoprotein heterodimer.Curr Issues Mol Biol. 2007 Jul;9(2):71-86. Curr Issues Mol Biol. 2007. PMID: 17489436 Review.
Cited by
-
Hepatitis C Virus Envelope Glycoproteins: A Balancing Act of Order and Disorder.Front Immunol. 2018 Aug 24;9:1917. doi: 10.3389/fimmu.2018.01917. eCollection 2018. Front Immunol. 2018. PMID: 30197646 Free PMC article. Review.
-
Designing a B Cell-Based Vaccine against a Highly Variable Hepatitis C Virus.Front Microbiol. 2018 Jan 15;8:2692. doi: 10.3389/fmicb.2017.02692. eCollection 2017. Front Microbiol. 2018. PMID: 29379486 Free PMC article. Review.
-
Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.Nat Commun. 2023 Jul 5;14(1):3980. doi: 10.1038/s41467-023-39659-z. Nat Commun. 2023. PMID: 37407593 Free PMC article.
-
A Biologically-validated HCV E1E2 Heterodimer Structural Model.Sci Rep. 2017 Mar 16;7(1):214. doi: 10.1038/s41598-017-00320-7. Sci Rep. 2017. PMID: 28303031 Free PMC article.
-
Computational Prediction of the Heterodimeric and Higher-Order Structure of gpE1/gpE2 Envelope Glycoproteins Encoded by Hepatitis C Virus.J Virol. 2017 Mar 29;91(8):e02309-16. doi: 10.1128/JVI.02309-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148799 Free PMC article.
References
-
- Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi:10.1128/JVI.74.8.3623-3633.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

