Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture
- PMID: 26246576
- PMCID: PMC4580162
- DOI: 10.1128/JVI.01411-15
Neuraminidase of Influenza A Virus Binds Lysosome-Associated Membrane Proteins Directly and Induces Lysosome Rupture
Abstract
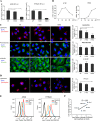
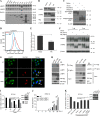
As a recycling center, lysosomes are filled with numerous acid hydrolase enzymes that break down waste materials and invading pathogens. Recently, lysosomal cell death has been defined as "lysosomal membrane permeabilization and the consequent leakage of lysosome contents into cytosol." Here, we show that the neuraminidase (NA) of H5N1 influenza A virus markedly deglycosylates and degrades lysosome-associated membrane proteins (LAMPs; the most abundant membrane proteins of lysosome), which induces lysosomal rupture, and finally leads to cell death of alveolar epithelial carcinoma A549 cells and human tracheal epithelial cells. The NA inhibitors peramivir and zanamivir could effectively block the deglycosylation of LAMPs, inhibit the virus cell entry, and prevent cell death induced by the H5N1 influenza virus. The NA of seasonal H1N1 virus, however, does not share these characteristics. Our findings not only reveal a novel role of NA in the early stage of the H5N1 influenza virus life cycle but also elucidate the molecular mechanism of lysosomal rupture crucial for influenza virus induced cell death.
Importance: The integrity of lysosomes is vital for maintaining cell homeostasis, cellular defense and clearance of invading pathogens. This study shows that the H5N1 influenza virus could induce lysosomal rupture through deglycosylating lysosome-associated membrane proteins (LAMPs) mediated by the neuraminidase activity of NA protein. NA inhibitors such as peramivir and zanamivir could inhibit the deglycosylation of LAMPs and protect lysosomes, which also further interferes with the H5N1 influenza virus infection at early stage of life cycle. This work is significant because it presents new concepts for NA's function, as well as for influenza inhibitors' mechanism of action, and could partially explain the high mortality and high viral load after H5N1 virus infection in human beings and why NA inhibitors have more potent therapeutic effects for lethal avian influenza virus infections at early stage.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Dynamic behavior of avian influenza A virus neuraminidase subtype H5N1 in complex with oseltamivir, zanamivir, peramivir, and their phosphonate analogues.J Chem Inf Model. 2009 Oct;49(10):2323-32. doi: 10.1021/ci900277r. J Chem Inf Model. 2009. PMID: 19780597
-
Recent advances in neuraminidase inhibitor development as anti-influenza drugs.ChemMedChem. 2012 Sep;7(9):1527-36. doi: 10.1002/cmdc.201200155. Epub 2012 Jul 16. ChemMedChem. 2012. PMID: 22807317 Review.
-
Neuraminidase inhibitor susceptibility profile of pandemic and seasonal influenza viruses during the 2009-2010 and 2010-2011 influenza seasons in Japan.Antiviral Res. 2013 Sep;99(3):261-9. doi: 10.1016/j.antiviral.2013.06.003. Epub 2013 Jun 17. Antiviral Res. 2013. PMID: 23791870
-
[Virological impact of stalk region of neuraminidase in influenza A/Anhui/1/05 (H5N1) and A/Ohio/07/2009 (H1N1) viruses].Bing Du Xue Bao. 2014 May;30(3):238-45. Bing Du Xue Bao. 2014. PMID: 25118377 Chinese.
-
Laninamivir octanoate: a new long-acting neuraminidase inhibitor for the treatment of influenza.Expert Rev Anti Infect Ther. 2011 Oct;9(10):851-7. doi: 10.1586/eri.11.112. Expert Rev Anti Infect Ther. 2011. PMID: 21973296 Review.
Cited by
-
Organelle dynamics and viral infections: at cross roads.Microbes Infect. 2019 Jan-Feb;21(1):20-32. doi: 10.1016/j.micinf.2018.06.002. Epub 2018 Jun 25. Microbes Infect. 2019. PMID: 29953921 Free PMC article. Review.
-
Zinc oxide nanoparticles harness autophagy to induce cell death in lung epithelial cells.Cell Death Dis. 2017 Jul 27;8(7):e2954. doi: 10.1038/cddis.2017.337. Cell Death Dis. 2017. PMID: 28749469 Free PMC article.
-
Ambient atmospheric PM worsens mouse lung injury induced by influenza A virus through lysosomal dysfunction.Respir Res. 2023 Dec 6;24(1):306. doi: 10.1186/s12931-023-02618-9. Respir Res. 2023. PMID: 38057804 Free PMC article.
-
The HN protein of Newcastle disease virus induces cell apoptosis through the induction of lysosomal membrane permeabilization.PLoS Pathog. 2024 Feb 14;20(2):e1011981. doi: 10.1371/journal.ppat.1011981. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38354122 Free PMC article.
-
Southern rice black-streaked dwarf virus induces incomplete autophagy for persistence in gut epithelial cells of its vector insect.PLoS Pathog. 2023 Jan 27;19(1):e1011134. doi: 10.1371/journal.ppat.1011134. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36706154 Free PMC article.
References
-
- Schneede A, Schmidt CK, Holtta-Vuori M, Heeren J, Willenborg M, Blanz J, Domanskyy M, Breiden B, Brodesser S, Landgrebe J, Sandhoff K, Ikonen E, Saftig P, Eskelinen EL. 2011. Role for LAMP-2 in endosomal cholesterol transport. J Cell Mol Med 15:280–295. doi:10.1111/j.1582-4934.2009.00973.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

