Phylogeny of Echinoderm Hemoglobins
- PMID: 26247465
- PMCID: PMC4527676
- DOI: 10.1371/journal.pone.0129668
Phylogeny of Echinoderm Hemoglobins
Abstract
Background: Recent genomic information has revealed that neuroglobin and cytoglobin are the two principal lineages of vertebrate hemoglobins, with the latter encompassing the familiar myoglobin and α-globin/β-globin tetramer hemoglobin, and several minor groups. In contrast, very little is known about hemoglobins in echinoderms, a phylum of exclusively marine organisms closely related to vertebrates, beyond the presence of coelomic hemoglobins in sea cucumbers and brittle stars. We identified about 50 hemoglobins in sea urchin, starfish and sea cucumber genomes and transcriptomes, and used Bayesian inference to carry out a molecular phylogenetic analysis of their relationship to vertebrate sequences, specifically, to assess the hypothesis that the neuroglobin and cytoglobin lineages are also present in echinoderms.
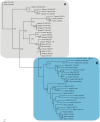
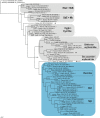
Results: The genome of the sea urchin Strongylocentrotus purpuratus encodes several hemoglobins, including a unique chimeric 14-domain globin, 2 androglobin isoforms and a unique single androglobin domain protein. Other strongylocentrotid genomes appear to have similar repertoires of globin genes. We carried out molecular phylogenetic analyses of 52 hemoglobins identified in sea urchin, brittle star and sea cucumber genomes and transcriptomes, using different multiple sequence alignment methods coupled with Bayesian and maximum likelihood approaches. The results demonstrate that there are two major globin lineages in echinoderms, which are related to the vertebrate neuroglobin and cytoglobin lineages. Furthermore, the brittle star and sea cucumber coelomic hemoglobins appear to have evolved independently from the cytoglobin lineage, similar to the evolution of erythroid oxygen binding globins in cyclostomes and vertebrates.
Conclusion: The presence of echinoderm globins related to the vertebrate neuroglobin and cytoglobin lineages suggests that the split between neuroglobins and cytoglobins occurred in the deuterostome ancestor shared by echinoderms and vertebrates.
Conflict of interest statement
Figures





References
-
- Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407(6803):520–3. - PubMed
-
- Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol. 2002;19(4):416–21. 11919282 - PubMed
-
- Kawada N, Kristensen DB, Asahina K, Nakatani K, Minamiyama Y, Seki S, et al. Characterization of a stellate cell activation-associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells. J Biol Chem. 2001;276(27):25318–23. - PubMed
-
- Trent JT 3rd, Hargrove MS. A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem. 2002;277(22):19538–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

