A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- PMID: 26247883
- PMCID: PMC4527830
- DOI: 10.1371/journal.pgen.1005429
A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
Abstract
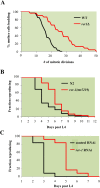
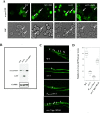
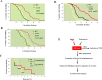
Proliferating eukaryotic cells undergo a finite number of cell divisions before irreversibly exiting mitosis. Yet pathways that normally limit the number of cell divisions remain poorly characterized. Here we describe a screen of a collection of 3762 single gene mutants in the yeast Saccharomyces cerevisiae, accounting for 2/3 of annotated yeast ORFs, to search for mutants that undergo an atypically high number of cell divisions. Many of the potential longevity genes map to cellular processes not previously implicated in mitotic senescence, suggesting that regulatory mechanisms governing mitotic exit may be broader than currently anticipated. We focused on an ER-Golgi gene cluster isolated in this screen to determine how these ubiquitous organelles integrate into mitotic longevity. We report that a chronic increase in ER protein load signals an expansion in the assembly of autophagosomes in an Ire1-independent manner, accelerates trafficking of high molecular weight protein aggregates from the cytoplasm to the vacuoles, and leads to a profound enhancement of daughter cell production. We demonstrate that this catabolic network is evolutionarily conserved, as it also extends reproductive lifespan in the nematode Caenorhabditis elegans. Our data provide evidence that catabolism of protein aggregates, a natural byproduct of high protein synthesis and turn over in dividing cells, is among the drivers of mitotic longevity in eukaryotes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hayflick L (1965) The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res 37: 614–636. - PubMed
-
- Smith JR, Pereira-Smith OM (1996) Replicative senescence: implications for in vivo aging and tumor suppression. Science 273: 63–67. - PubMed
-
- Jazwinski SM, Egilmez NK, Chen JB (1989) Replication control and cellular life span. Exp Gerontol 24: 423–436. - PubMed
-
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, et al. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

