Fluconazole and Voriconazole Resistance in Candida parapsilosis Is Conferred by Gain-of-Function Mutations in MRR1 Transcription Factor Gene
- PMID: 26248365
- PMCID: PMC4576118
- DOI: 10.1128/AAC.00842-15
Fluconazole and Voriconazole Resistance in Candida parapsilosis Is Conferred by Gain-of-Function Mutations in MRR1 Transcription Factor Gene
Abstract
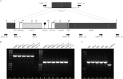
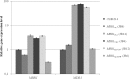
Candida parapsilosis is the second most prevalent fungal agent causing bloodstream infections. Nevertheless, there is little information about the molecular mechanisms underlying azole resistance in this species. Mutations (G1747A, A2619C, and A3191C) in the MRR1 transcription factor gene were identified in fluconazole- and voriconazole-resistant strains. Independent expression of MRR1 genes harboring these mutations showed that G1747A (G583R) and A2619C (K873N) are gain-of-function mutations responsible for azole resistance, the first described in C. parapsilosis.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Montagna MT, Lovero G, Borghi E, Amato G, Andreoni S, Campion L, Lo Cascio G, Lombardi G, Luzzaro F, Manso E, Mussap M, Pecile P, Perin S, Tangorra E, Tronci M, Iatta R, Morace G. 2014. Candidemia in intensive care unit: a nationwide prospective observational survey (GISIA-3 study) and review of the European literature from 2000 through 2013. Eur Rev Med Pharmacol Sci 18:661–674. - PubMed
-
- Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob Agents Chemother 52:2305–2312. doi:10.1128/AAC.00262-08. - DOI - PMC - PubMed
-
- Klingspor L, Tortorano AM, Peman J, Willinger B, Hamal P, Sendid B, Velegraki A, Kibbler C, Meis JF, Sabino R, Ruhnke M, Arikan-Akdagli S, Salonen J, Doczi I. 2015. Invasive Candida infections in surgical patients in intensive care units: a prospective, multicentre survey initiated by the European Confederation of Medical Mycology (ECMM) (2006-2008). Clin Microbiol Infect 21:87.e1–87.e10. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

