Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster
- PMID: 26248466
- PMCID: PMC4528719
- DOI: 10.1186/s12915-015-0168-7
Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster
Abstract
Background: Insulators play a central role in gene regulation, chromosomal architecture and genome function in higher eukaryotes. To learn more about how insulators carry out their diverse functions, we have begun an analysis of the Drosophila CTCF (dCTCF). CTCF is one of the few insulator proteins known to be conserved from flies to man.
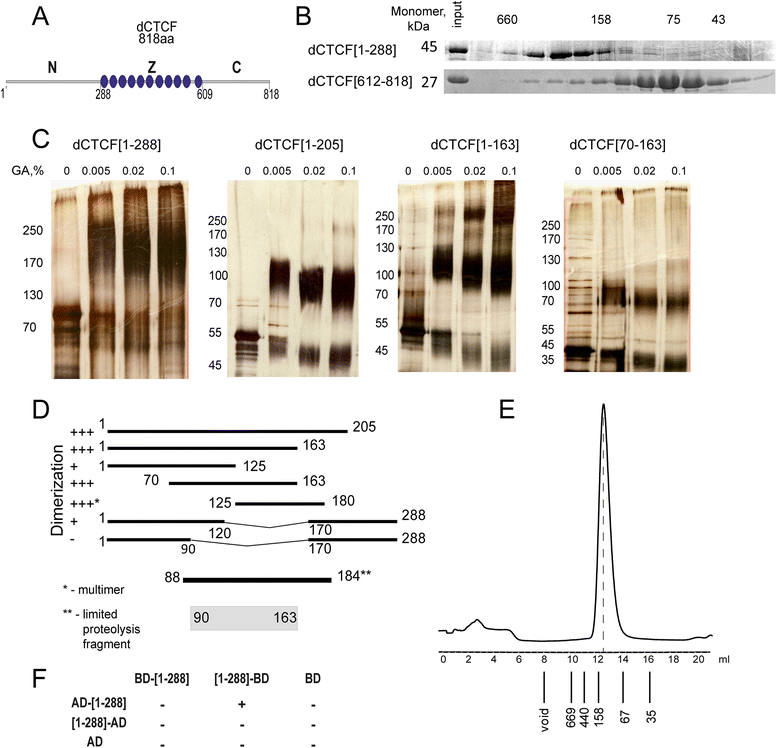
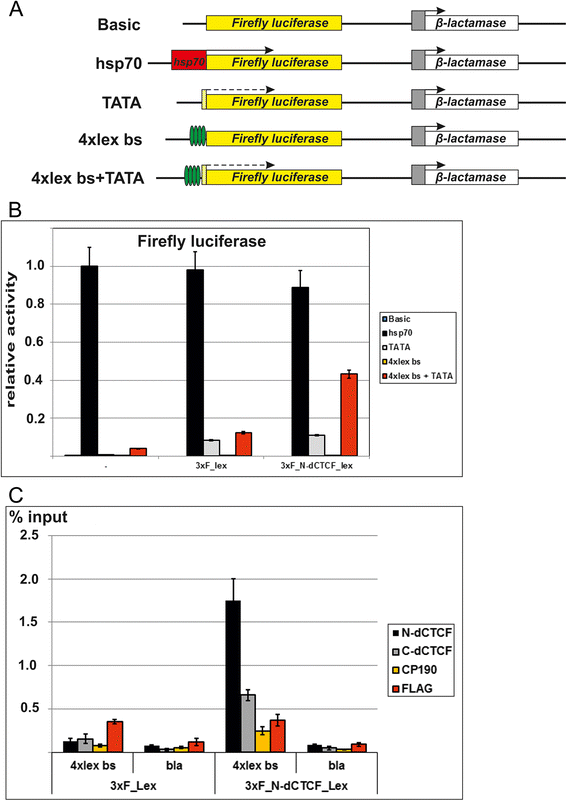
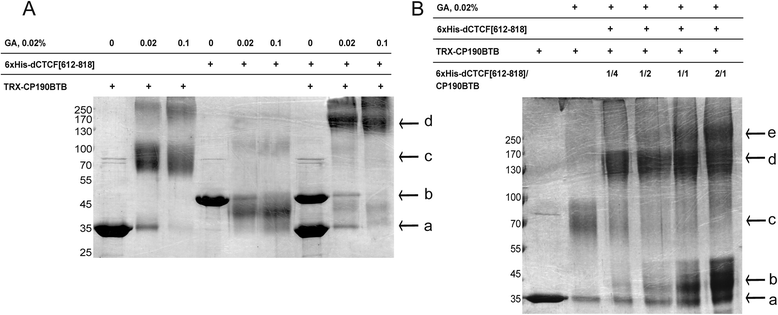
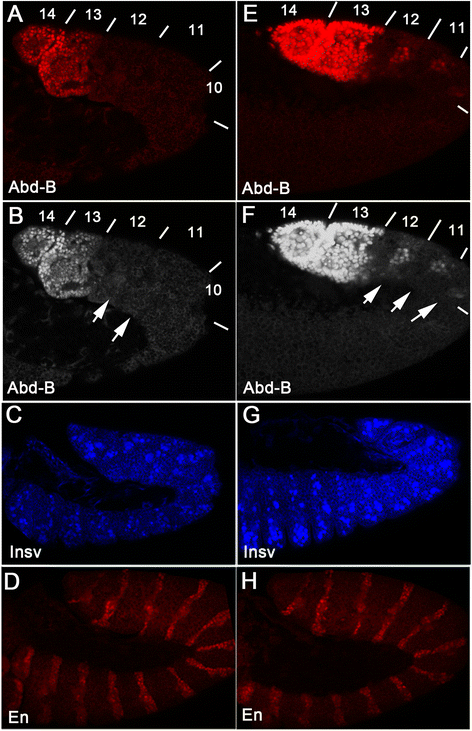
Results: In the studies reported here we have focused on the identification and characterization of two dCTCF protein interaction modules. The first mediates dCTCF multimerization, while the second mediates dCTCF-CP190 interactions. The multimerization domain maps in the N-terminus of the dCTCF protein and likely mediates the formation of tetrameric complexes. The CP190 interaction module encompasses a sequence ~200 amino acids long that spans the C-terminal and mediates interactions with the N-terminal BTB domain of the CP190 protein. Transgene rescue experiments showed that a dCTCF protein lacking sequences critical for CP190 interactions was almost as effective as wild type in rescuing the phenotypic effects of a dCTCF null allele. The mutation did, however, affect CP190 recruitment to specific Drosophila insulator elements and had a modest effect on dCTCF chromatin association. A protein lacking the N-terminal dCTCF multimerization domain incompletely rescued the zygotic and maternal effect lethality of the null and did not rescue the defects in Abd-B regulation evident in surviving adult dCTCF mutant flies. Finally, we show that elimination of maternally contributed dCTCF at the onset of embryogenesis has quite different effects on development and Abd-B regulation than is observed when the homozygous mutant animals develop in the presence of maternally derived dCTCF activity.
Conclusions: Our results indicate that dCTCF-CP190 interactions are less critical for the in vivo functions of the dCTCF protein than the N-terminal dCTCF-dCTCF interaction domain. We also show that the phenotypic consequences of dCTCF mutations differ depending upon when and how dCTCF activity is lost.
Figures





Comment in
-
In vivo studies of the Drosophila insulator factor CTCF reach a Catch 22.BMC Biol. 2015 Sep 2;13:71. doi: 10.1186/s12915-015-0182-9. BMC Biol. 2015. PMID: 26329471 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

