Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy
- PMID: 26248552
- PMCID: PMC4527111
- DOI: 10.1186/s12916-015-0421-5
Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy
Abstract
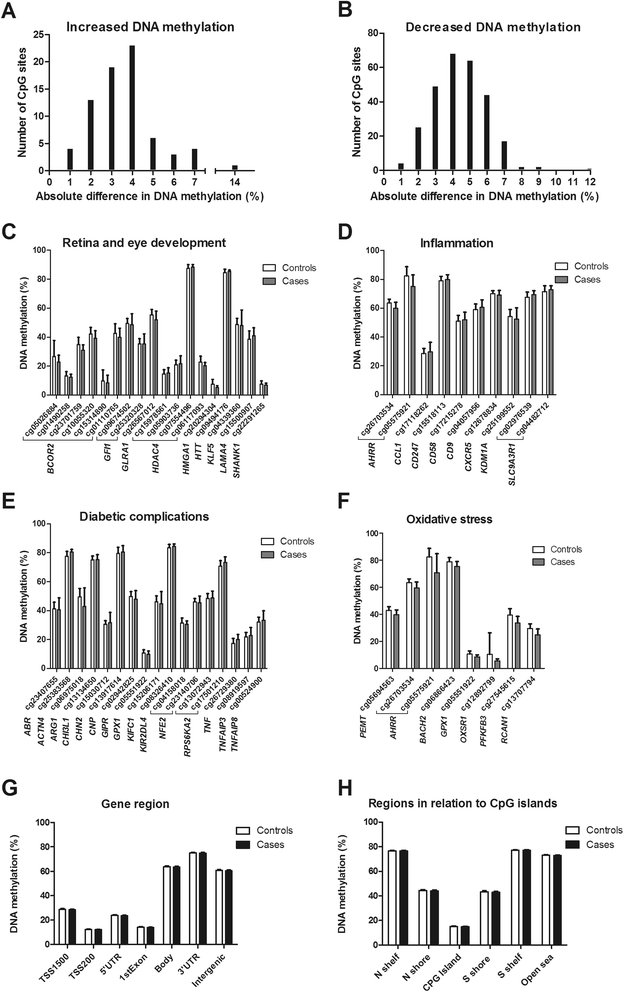
Background: Epigenetic variation has been linked to several human diseases. Proliferative diabetic retinopathy (PDR) is a major cause of vision loss in subjects with diabetes. However, studies examining the association between PDR and the genome-wide DNA methylation pattern are lacking. Our aim was to identify epigenetic modifications that associate with and predict PDR in subjects with type 1 diabetes (T1D).
Methods: DNA methylation was analyzed genome-wide in 485,577 sites in blood from cases with PDR (n = 28), controls (n = 30), and in a prospective cohort (n = 7). False discovery rate analysis was used to correct the data for multiple testing. Study participants with T1D diagnosed before 30 years of age and insulin treatment within 1 year from diagnosis were selected based on 1) subjects classified as having PDR (cases) and 2) subjects with T1D who had had diabetes for at least 10 years when blood DNA was sampled and classified as having no/mild diabetic retinopathy also after an 8.7-year follow-up (controls). DNA methylation was also analyzed in a prospective cohort including seven subjects with T1D who had no/mild diabetic retinopathy when blood samples were taken, but who developed PDR within 6.3 years (converters). The retinopathy level was classified by fundus photography.
Results: We identified differential DNA methylation of 349 CpG sites representing 233 unique genes including TNF, CHI3L1 (also known as YKL-40), CHN2, GIPR, GLRA1, GPX1, AHRR, and BCOR in cases with PDR compared with controls. The majority of these sites (79 %) showed decreased DNA methylation in cases with PDR. The Natural Killer cell-mediated cytotoxicity pathway was found to be significantly (P = 0.006) enriched among differentially methylated genes in cases with PDR. We also identified differential DNA methylation of 28 CpG sites representing 17 genes (e.g. AHRR, GIPR, GLRA1, and BCOR) with P <0.05 in the prospective cohort, which is more than expected by chance (P = 0.0096).
Conclusions: Subjects with T1D and PDR exhibit altered DNA methylation patterns in blood. Some of these epigenetic changes may predict the development of PDR, suggesting that DNA methylation may be used as a prospective marker of PDR.
Figures
Similar articles
-
Genome-wide analysis of DNA methylation identifies S100A13 as an epigenetic biomarker in individuals with chronic (≥ 30 years) type 2 diabetes without diabetic retinopathy.Clin Epigenetics. 2020 Jun 3;12(1):77. doi: 10.1186/s13148-020-00871-z. Clin Epigenetics. 2020. PMID: 32493412 Free PMC article.
-
Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis.PLoS Genet. 2011 Sep;7(9):e1002300. doi: 10.1371/journal.pgen.1002300. Epub 2011 Sep 29. PLoS Genet. 2011. PMID: 21980303 Free PMC article.
-
An Epigenome-Wide Association Study of DNA Methylation and Proliferative Retinopathy over 28 Years in Type 1 Diabetes.Ophthalmol Sci. 2024 Feb 28;4(4):100497. doi: 10.1016/j.xops.2024.100497. eCollection 2024 Jul-Aug. Ophthalmol Sci. 2024. PMID: 38601260 Free PMC article.
-
Novel epigenetic-sensitive clinical challenges both in type 1 and type 2 diabetes.J Diabetes Complications. 2018 Nov;32(11):1076-1084. doi: 10.1016/j.jdiacomp.2018.08.012. Epub 2018 Aug 19. J Diabetes Complications. 2018. PMID: 30190170 Review.
-
Role of histone modification and DNA methylation in signaling pathways involved in diabetic retinopathy.J Cell Physiol. 2019 Jun;234(6):7839-7846. doi: 10.1002/jcp.27844. Epub 2018 Dec 4. J Cell Physiol. 2019. PMID: 30515789 Review.
Cited by
-
Obesity-Related Epigenetic Changes After Bariatric Surgery.Front Endocrinol (Lausanne). 2019 Apr 16;10:232. doi: 10.3389/fendo.2019.00232. eCollection 2019. Front Endocrinol (Lausanne). 2019. PMID: 31040824 Free PMC article. Review.
-
Epigenetic Changes Induced by Maternal Factors during Fetal Life: Implication for Type 1 Diabetes.Genes (Basel). 2021 Jun 8;12(6):887. doi: 10.3390/genes12060887. Genes (Basel). 2021. PMID: 34201206 Free PMC article. Review.
-
The Role of Epigenetic Modifications in Late Complications in Type 1 Diabetes.Genes (Basel). 2022 Apr 15;13(4):705. doi: 10.3390/genes13040705. Genes (Basel). 2022. PMID: 35456511 Free PMC article. Review.
-
DNA methyltransferase expression (DNMT1, DNMT3a and DNMT3b) as a potential biomarker for anti-VEGF diabetic macular edema response.Eur J Ophthalmol. 2023 Nov;33(6):2267-2274. doi: 10.1177/11206721231171623. Epub 2023 Apr 20. Eur J Ophthalmol. 2023. PMID: 37082811 Free PMC article.
-
Role of Epigenetics in Type 2 Diabetes and Obesity.Biomedicines. 2021 Aug 8;9(8):977. doi: 10.3390/biomedicines9080977. Biomedicines. 2021. PMID: 34440181 Free PMC article. Review.
References
-
- White NH, Sun W, Cleary PA, Danis RP, Davis MD, Hainsworth DP, et al. Prolonged effect of intensive therapy on the risk of retinopathy complications in patients with type 1 diabetes mellitus: 10 years after the Diabetes Control and Complications Trial. Arch Ophthalmol. 2008;126:1707–1715. doi: 10.1001/archopht.126.12.1707. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

