Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics
- PMID: 26248648
- PMCID: PMC4828339
- DOI: 10.1158/1541-7786.MCR-15-0189
Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics
Abstract
Compared with normal cells, tumor cells have undergone an array of genetic and epigenetic alterations. Often, these changes underlie cancer development, progression, and drug resistance, so the utility of model systems rests on their ability to recapitulate the genomic aberrations observed in primary tumors. Tumor-derived cell lines have long been used to study the underlying biologic processes in cancer, as well as screening platforms for discovering and evaluating the efficacy of anticancer therapeutics. Multiple -omic measurements across more than a thousand cancer cell lines have been produced following advances in high-throughput technologies and multigroup collaborative projects. These data complement the large, international cancer genomic sequencing efforts to characterize patient tumors, such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). Given the scope and scale of data that have been generated, researchers are now in a position to evaluate the similarities and differences that exist in genomic features between cell lines and patient samples. As pharmacogenomics models, cell lines offer the advantages of being easily grown, relatively inexpensive, and amenable to high-throughput testing of therapeutic agents. Data generated from cell lines can then be used to link cellular drug response to genomic features, where the ultimate goal is to build predictive signatures of patient outcome. This review highlights the recent work that has compared -omic profiles of cell lines with primary tumors, and discusses the advantages and disadvantages of cancer cell lines as pharmacogenomic models of anticancer therapies.
©2015 American Association for Cancer Research.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures
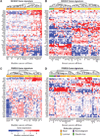
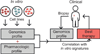
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

