Transplant biopsy beyond light microscopy
- PMID: 26249165
- PMCID: PMC4528359
- DOI: 10.1186/s12882-015-0136-z
Transplant biopsy beyond light microscopy
Abstract
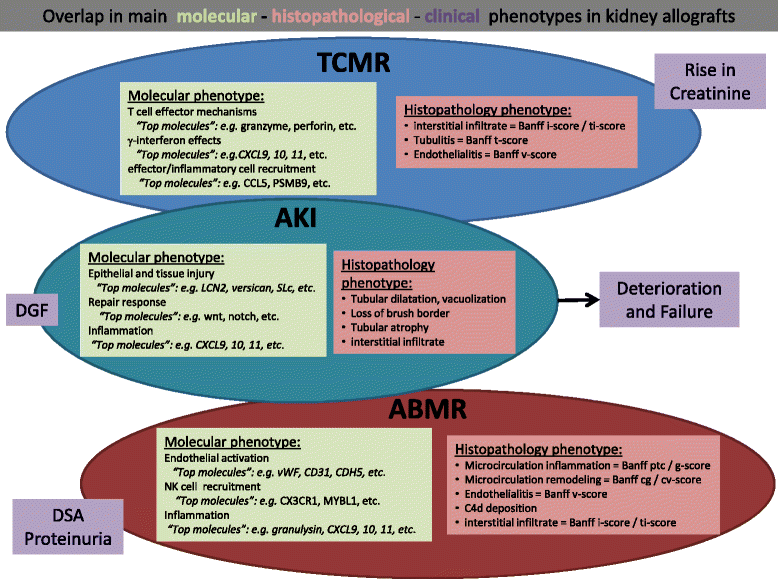
Despite its long-standing status as the diagnostic "gold standard", the renal transplant biopsy is limited by a fundamental dependence on descriptive, empirically-derived consensus classification. The recent shift towards personalized medicine has resulted in an increased demand for precise, mechanism-based diagnoses, which is not fully met by the contemporary transplantation pathology standard of care. The expectation is that molecular techniques will provide novel pathogenetic insights that will allow for the identification of more accurate diagnostic, prognostic, and therapeutic targets. Here we review the current state of molecular renal transplantation pathology. Despite significant research activity and progress within the field, routine adoption of clinical molecular testing has not yet been achieved. The recent development of novel molecular platforms suitable for use with formalin-fixed paraffin-embedded tissue will offer potential solution for the major barriers to implementation. The recent incorporation of molecular diagnostic criteria into the 2013 Banff classification is a reflection of progress made and future directions in the area of molecular transplantation pathology. Transcripts related to endothelial injury and NK cell activation have consistently been shown to be associated with antibody-mediated rejection. Prospective multicenter validation and implementation of molecular diagnostics for major entities remains an unmet clinical need in transplantation. It is expected that an integrated system of transplantation pathology diagnosis comprising molecular, morphological, serological, and clinical variables will ultimately provide the greatest diagnostic precision.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

