Growth differentiation factor-15: a p53- and demethylation-upregulating gene represses cell proliferation, invasion, and tumorigenesis in bladder carcinoma cells
- PMID: 26249737
- PMCID: PMC4528199
- DOI: 10.1038/srep12870
Growth differentiation factor-15: a p53- and demethylation-upregulating gene represses cell proliferation, invasion, and tumorigenesis in bladder carcinoma cells
Abstract
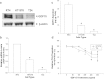
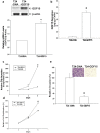
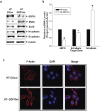
Growth differentiation factor-15 (GDF15), a member of the TGF-β superfamily, affects tumor biology of certain cancers, but remains poorly understood in bladder cancer cells. This study determined the expression, regulation, function, and potential downstream target genes of GDF15 in bladder carcinoma cells. The transitional papilloma carcionoma cells (RT4) expressed higher levels of GDF15 as compared with the bladder carcinoma cells (HT1376 and T24). Treatments of recombinant human GDF15 (rhGDF15) reduced the proliferations of HT1376 and T24 cells. Expression of GDF15 was upregulated via DNA demethylation and p53. The cell proliferation, invasion, and tumorigenesis were reduced in ectopic overexpression of GDF15, while enhanced in GDF15 knockdown. The expressions of mammary serine protease inhibitor (MASPIN) and N-myc downstream-regulated family genes (NDRG1, NDRG2, and NDRG3) were upregulated by GDF15 overexpressions and rhGDF15 treatments in bladder carcinoma cells. GDF15 knockdown induced epithelial-mesenchymal transition (EMT) and F-actin polarization in HT1376 cells. Our results suggest that enhanced expressions of MASPIN and N-myc downstream-regulated family genes and the modulation of EMT may account for the inhibitory functions of GDF15 in the cell proliferation, invasion, and tumorigenesis of bladder carcinoma cells. The GDF15 should be considered as a tumor suppressor in human bladder carcinoma cells.
Figures




Similar articles
-
Metallothionein 3 Is a Hypoxia-Upregulated Oncogene Enhancing Cell Invasion and Tumorigenesis in Human Bladder Carcinoma Cells.Int J Mol Sci. 2019 Feb 23;20(4):980. doi: 10.3390/ijms20040980. Int J Mol Sci. 2019. PMID: 30813460 Free PMC article.
-
Prostate-derived ets factor represses tumorigenesis and modulates epithelial-to-mesenchymal transition in bladder carcinoma cells.Cancer Lett. 2016 May 28;375(1):142-151. doi: 10.1016/j.canlet.2016.02.056. Epub 2016 Mar 7. Cancer Lett. 2016. PMID: 26965996
-
Growth differentiation factor-15 upregulates interleukin-6 to promote tumorigenesis of prostate carcinoma PC-3 cells.J Mol Endocrinol. 2012 Sep 5;49(2):153-63. doi: 10.1530/JME-11-0149. Print 2012 Oct. J Mol Endocrinol. 2012. PMID: 22872134
-
The p53 tumor suppressor gene and nuclear protein: basic science review and relevance in the management of bladder cancer.J Urol. 2003 Apr;169(4):1219-28. doi: 10.1097/01.ju.0000056085.58221.80. J Urol. 2003. PMID: 12629332 Review.
-
[Tumorigenesis from a pathological perspective : Tumor spread and epigenetically regulated genes in bladder cancer].Pathologe. 2016 Nov;37(Suppl 2):196-203. doi: 10.1007/s00292-016-0207-3. Pathologe. 2016. PMID: 27613302 Review. German.
Cited by
-
Macrophages as a Source and Target of GDF-15.Int J Mol Sci. 2024 Jul 3;25(13):7313. doi: 10.3390/ijms25137313. Int J Mol Sci. 2024. PMID: 39000420 Free PMC article. Review.
-
The Differential Antitumor Activity of 5-Aza-2'-deoxycytidine in Prostate Cancer DU145, 22RV1, and LNCaP Cells.J Cancer. 2021 Jul 25;12(18):5593-5604. doi: 10.7150/jca.56709. eCollection 2021. J Cancer. 2021. PMID: 34405020 Free PMC article.
-
Activating transcription factor 3 is an antitumor gene synergizing with growth differentiation factor 15 to modulate cell growth in human bladder cancer.Biomed J. 2025 Apr;48(2):100756. doi: 10.1016/j.bj.2024.100756. Epub 2024 Jun 27. Biomed J. 2025. PMID: 38942385 Free PMC article.
-
The prognostic value of six survival-related genes in bladder cancer.Cell Death Discov. 2020 Jul 13;6:58. doi: 10.1038/s41420-020-00295-x. eCollection 2020. Cell Death Discov. 2020. PMID: 32695477 Free PMC article.
-
Metallothionein 3 Is a Hypoxia-Upregulated Oncogene Enhancing Cell Invasion and Tumorigenesis in Human Bladder Carcinoma Cells.Int J Mol Sci. 2019 Feb 23;20(4):980. doi: 10.3390/ijms20040980. Int J Mol Sci. 2019. PMID: 30813460 Free PMC article.
References
-
- Siegel R., Naishadham D. & Jemal A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30 (2013). - PubMed
-
- Mitra A. P. & Cota R. J. Molecular pathogenesis and diagnostics of bladder cancer. Annu Rev Pathol 4, 251–285 (2009). - PubMed
-
- Ghafouri-Fard S., Nekoohesh L. & Motevaseli E. Bladder cancer biomarkers: Review and update. Asian Pac J Cancer Prev 15, 2395–2403 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

