Global diversity and biogeography of deep-sea pelagic prokaryotes
- PMID: 26251871
- PMCID: PMC4817678
- DOI: 10.1038/ismej.2015.137
Global diversity and biogeography of deep-sea pelagic prokaryotes
Abstract
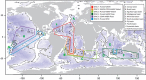
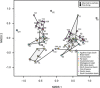
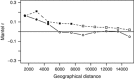
The deep-sea is the largest biome of the biosphere, and contains more than half of the whole ocean's microbes. Uncovering their general patterns of diversity and community structure at a global scale remains a great challenge, as only fragmentary information of deep-sea microbial diversity exists based on regional-scale studies. Here we report the first globally comprehensive survey of the prokaryotic communities inhabiting the bathypelagic ocean using high-throughput sequencing of the 16S rRNA gene. This work identifies the dominant prokaryotes in the pelagic deep ocean and reveals that 50% of the operational taxonomic units (OTUs) belong to previously unknown prokaryotic taxa, most of which are rare and appear in just a few samples. We show that whereas the local richness of communities is comparable to that observed in previous regional studies, the global pool of prokaryotic taxa detected is modest (~3600 OTUs), as a high proportion of OTUs are shared among samples. The water masses appear to act as clear drivers of the geographical distribution of both particle-attached and free-living prokaryotes. In addition, we show that the deep-oceanic basins in which the bathypelagic realm is divided contain different particle-attached (but not free-living) microbial communities. The combination of the aging of the water masses and a lack of complete dispersal are identified as the main drivers for this biogeographical pattern. All together, we identify the potential of the deep ocean as a reservoir of still unknown biological diversity with a higher degree of spatial complexity than hitherto considered.
Figures



References
-
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430: 551–554. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

