More Cercospora Species Infect Soybeans across the Americas than Meets the Eye
- PMID: 26252018
- PMCID: PMC4529236
- DOI: 10.1371/journal.pone.0133495
More Cercospora Species Infect Soybeans across the Americas than Meets the Eye
Abstract
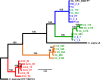
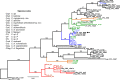
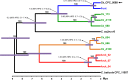
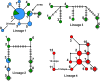
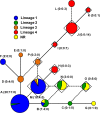
Diseases of soybean caused by Cercospora spp. are endemic throughout the world's soybean production regions. Species diversity in the genus Cercospora has been underestimated due to overdependence on morphological characteristics, symptoms, and host associations. Currently, only two species (Cercospora kikuchii and C. sojina) are recognized to infect soybean; C. kikuchii causes Cercospora leaf blight (CLB) and purple seed stain (PSS), whereas C. sojina causes frogeye leaf spot. To assess cryptic speciation among pathogens causing CLB and PSS, phylogenetic and phylogeographic analyses were performed with isolates from the top three soybean producing countries (USA, Brazil, and Argentina; collectively accounting for ~80% of global production). Eight nuclear genes and one mitochondrial gene were partially sequenced and analyzed. Additionally, amino acid substitutions conferring fungicide resistance were surveyed, and the production of cercosporin (a polyketide toxin produced by many Cercospora spp.) was assessed. From these analyses, the long-held assumption of C. kikuchii as the single causal agent of CLB and PSS was rejected experimentally. Four cercosporin-producing lineages were uncovered with origins (about 1 Mya) predicted to predate agriculture. Some of the Cercospora spp. newly associated with CLB and PSS appear to represent undescribed species; others were not previously reported to infect soybeans. Lineage 1, which contained the ex-type strain of C. kikuchii, was monophyletic and occurred in Argentina and Brazil. In contrast, lineages 2 and 3 were polyphyletic and contained wide-host range species complexes. Lineage 4 was monophyletic, thrived in Argentina and the USA, and included the generalist Cercospora cf. flagellaris. Interlineage recombination was detected, along with a high frequency of mutations linked to fungicide resistance in lineages 2 and 3. These findings point to cryptic Cercospora species as underappreciated global considerations for soybean production and phytosanitary vigilance, and urge a reassessment of host-specificity as a diagnostic tool for Cercospora.
Conflict of interest statement
Figures
Similar articles
-
Cercospora cf. flagellaris and Cercospora cf. sigesbeckiae Are Associated with Cercospora Leaf Blight and Purple Seed Stain on Soybean in North America.Phytopathology. 2016 Nov;106(11):1376-1385. doi: 10.1094/PHYTO-12-15-0332-R. Epub 2016 Aug 18. Phytopathology. 2016. PMID: 27183302
-
Distribution and Sequestration of Cercosporin by Cercospora cf. flagellaris.Phytopathology. 2024 Aug;114(8):1822-1831. doi: 10.1094/PHYTO-09-23-0310-R. Epub 2024 Aug 1. Phytopathology. 2024. PMID: 38700938
-
The AVR4 effector is involved in cercosporin biosynthesis and likely affects the virulence of Cercospora cf. flagellaris on soybean.Mol Plant Pathol. 2020 Jan;21(1):53-65. doi: 10.1111/mpp.12879. Epub 2019 Oct 23. Mol Plant Pathol. 2020. PMID: 31642594 Free PMC article.
-
Characterization of QoI-Fungicide Resistance in Cercospora Isolates Associated with Cercospora Leaf Blight of Soybean from the Southern United States.Plant Dis. 2024 Jan;108(1):149-161. doi: 10.1094/PDIS-03-23-0588-RE. Epub 2024 Jan 23. Plant Dis. 2024. PMID: 37578368
-
Phytopathogenic Cercosporoid Fungi-From Taxonomy to Modern Biochemistry and Molecular Biology.Int J Mol Sci. 2020 Nov 13;21(22):8555. doi: 10.3390/ijms21228555. Int J Mol Sci. 2020. PMID: 33202799 Free PMC article. Review.
Cited by
-
Culturable Endophytes Associated with Soybean Seeds and Their Potential for Suppressing Seed-Borne Pathogens.Plant Pathol J. 2022 Aug;38(4):313-322. doi: 10.5423/PPJ.OA.05.2022.0064. Epub 2022 Aug 1. Plant Pathol J. 2022. PMID: 35953051 Free PMC article.
-
High-quality genome assembly of the soybean fungal pathogen Cercospora kikuchii.G3 (Bethesda). 2021 Sep 27;11(10):jkab277. doi: 10.1093/g3journal/jkab277. G3 (Bethesda). 2021. PMID: 34568928 Free PMC article.
-
Molecular Breeding to Overcome Biotic Stresses in Soybean: Update.Plants (Basel). 2022 Jul 28;11(15):1967. doi: 10.3390/plants11151967. Plants (Basel). 2022. PMID: 35956444 Free PMC article. Review.
-
Deciphering genetic factors contributing to enhanced resistance against Cercospora leaf blight in soybean (Glycine max L.) using GWAS analysis.Front Genet. 2024 May 10;15:1377223. doi: 10.3389/fgene.2024.1377223. eCollection 2024. Front Genet. 2024. PMID: 38798696 Free PMC article.
-
Genome-wide association analysis of resistance to frogeye leaf spot China race 7 in soybean based on high-throughput sequencing.Theor Appl Genet. 2023 Apr 27;136(5):119. doi: 10.1007/s00122-023-04359-1. Theor Appl Genet. 2023. PMID: 37103627
References
-
- Pollack FG. An annotated compilation of Cercospora names. Mycol Mem. 1987;12: 1–212.
-
- Crous PW, Braun U. Mycosphaerella and its anamorphs. 1. Names published in Cercospora and Passalora . CBS Biodiversity Series 2003;1: 1–571.
-
- Fuckel KWGL. Fungi Rhenani exsiccati, Fasc. I-IV. Hedwigia 1863;2: 132–136.
-
- Chupp C. A monograph of the fungus genus Cercospora New York: The Ronald Press; 1954.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

