Methods for Improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool
- PMID: 26252519
- PMCID: PMC4529225
- DOI: 10.1371/journal.pone.0134802
Methods for Improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool
Erratum in
-
Correction: Methods for Improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool.PLoS One. 2015 Sep 25;10(9):e0139529. doi: 10.1371/journal.pone.0139529. eCollection 2015. PLoS One. 2015. PMID: 26406872 Free PMC article. No abstract available.
Abstract
Gut microbiome community analysis is used to understand many diseases like inflammatory bowel disease, obesity, and diabetes. Sampling methods are an important consideration for human microbiome research, yet are not emphasized in many studies. In this study, we demonstrate that the preparation, handling, and storage of human faeces are critical processes that alter the outcomes of downstream DNA-based bacterial community analyses via qPCR. We found that stool subsampling resulted in large variability of gut microbiome data due to different microenvironments harbouring various taxa within an individual stool. However, we reduced intra-sample variability by homogenizing the entire stool sample in liquid nitrogen and subsampling from the resulting crushed powder prior to DNA extraction. We experimentally determined that the bacterial taxa varied with room temperature storage beyond 15 minutes and beyond three days storage in a domestic frost-free freezer. While freeze thawing only had an effect on bacterial taxa abundance beyond four cycles, the use of samples stored in RNAlater should be avoided as overall DNA yields were reduced as well as the detection of bacterial taxa. Overall we provide solutions for processing and storing human stool samples that reduce variability of microbiome data. We recommend that stool is frozen within 15 minutes of being defecated, stored in a domestic frost-free freezer for less than three days, and homogenized prior to DNA extraction. Adoption of these simple protocols will have a significant and positive impact on future human microbiome research.
Conflict of interest statement
Figures
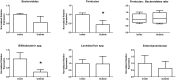


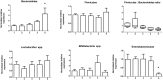


References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362: 709–715. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

