Adaptive mutations in sugar metabolism restore growth on glucose in a pyruvate decarboxylase negative yeast strain
- PMID: 26253003
- PMCID: PMC4529725
- DOI: 10.1186/s12934-015-0305-6
Adaptive mutations in sugar metabolism restore growth on glucose in a pyruvate decarboxylase negative yeast strain
Abstract
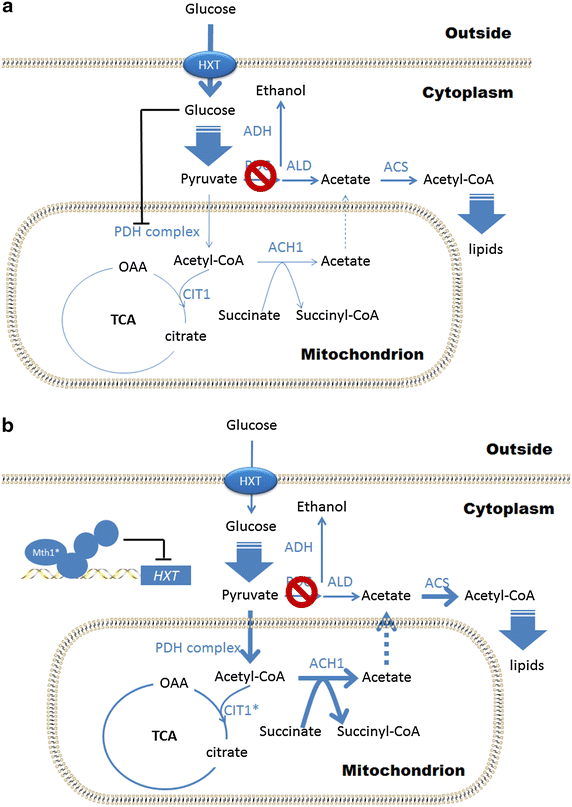
Background: A Saccharomyces cerevisiae strain carrying deletions in all three pyruvate decarboxylase (PDC) genes (also called Pdc negative yeast) represents a non-ethanol producing platform strain for the production of pyruvate derived biochemicals. However, it cannot grow on glucose as the sole carbon source, and requires supplementation of C2 compounds to the medium in order to meet the requirement for cytosolic acetyl-CoA for biosynthesis of fatty acids and ergosterol.
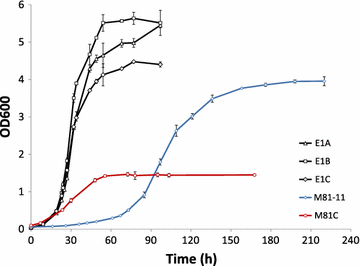
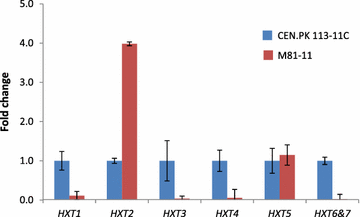
Results: In this study, a Pdc negative strain was adaptively evolved for improved growth in glucose medium via serial transfer, resulting in three independently evolved strains, which were able to grow in minimal medium containing glucose as the sole carbon source at the maximum specific rates of 0.138, 0.148, 0.141 h(-1), respectively. Several genetic changes were identified in the evolved Pdc negative strains by genomic DNA sequencing. Among these genetic changes, 4 genes were found to carry point mutations in at least two of the evolved strains: MTH1 encoding a negative regulator of the glucose-sensing signal transduction pathway, HXT2 encoding a hexose transporter, CIT1 encoding a mitochondrial citrate synthase, and RPD3 encoding a histone deacetylase. Reverse engineering of the non-evolved Pdc negative strain through introduction of the MTH1 (81D) allele restored its growth on glucose at a maximum specific rate of 0.053 h(-1) in minimal medium with 2% glucose, and the CIT1 deletion in the reverse engineered strain further increased the maximum specific growth rate to 0.069 h(-1).
Conclusions: In this study, possible evolving mechanisms of Pdc negative strains on glucose were investigated by genome sequencing and reverse engineering. The non-synonymous mutations in MTH1 alleviated the glucose repression by repressing expression of several hexose transporter genes. The non-synonymous mutations in HXT2 and CIT1 may function in the presence of mutated MTH1 alleles and could be related to an altered central carbon metabolism in order to ensure production of cytosolic acetyl-CoA in the Pdc negative strain.
Figures

References
-
- Jensen MK, Keasling JD. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2014 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

