M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide
- PMID: 26253167
- PMCID: PMC4545815
- DOI: 10.1186/s12885-015-1546-9
M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide
Abstract
Background: Tumor associated macrophages (TAMs) are present in high density in solid tumors. TAMs share many characteristics with alternatively activated macrophages, also called M2. They have been shown to favor tumor development and a role in chemoresistance has also been suggested. Here, we investigated the effects of M2 in comparison to M1 macrophages on cancer cell sensitivity to etoposide.
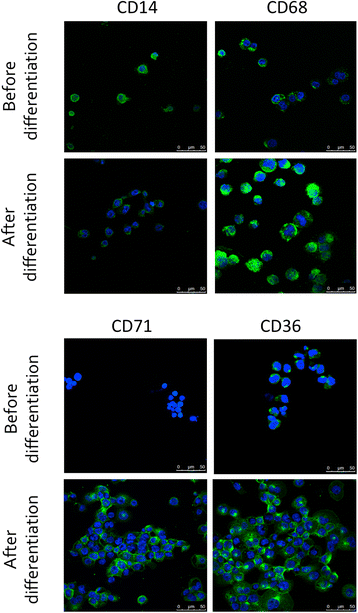
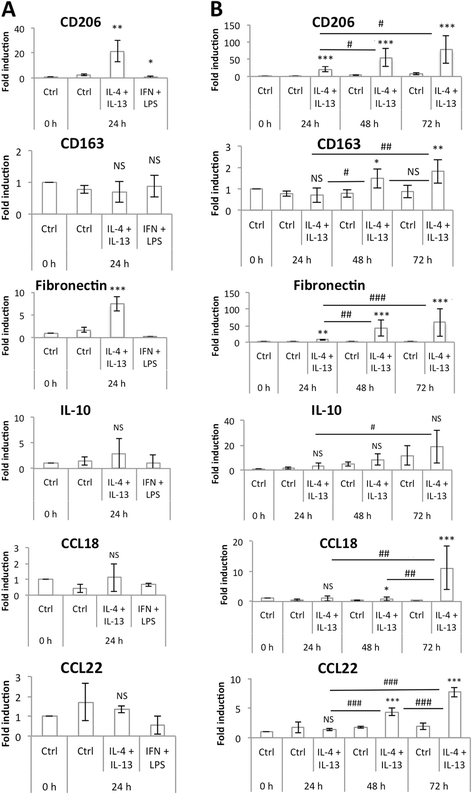
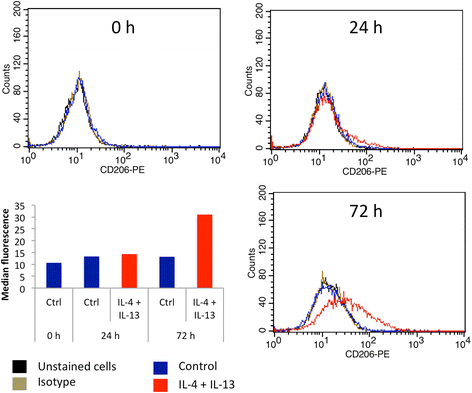
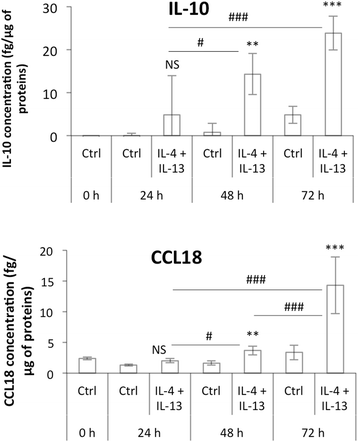
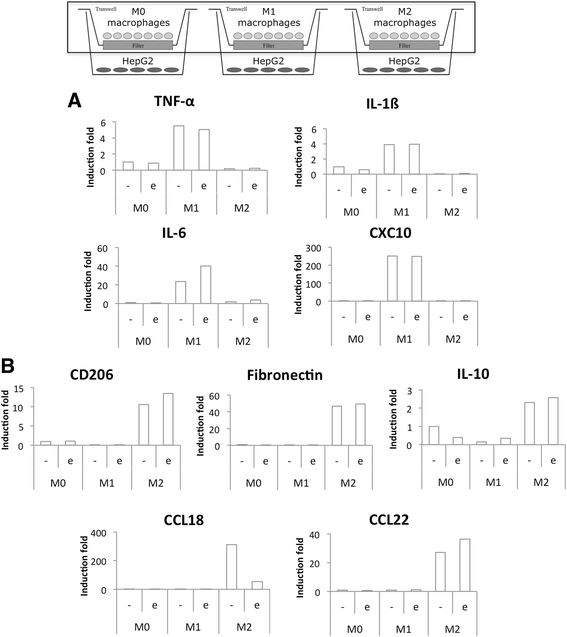
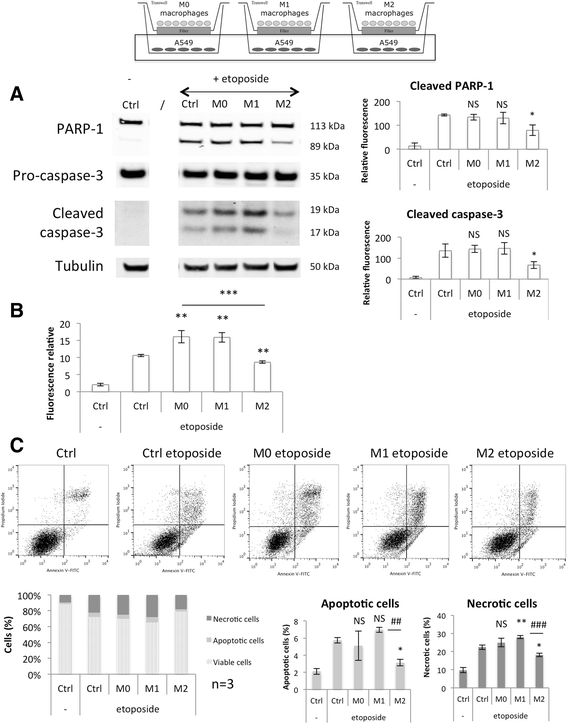
Methods: We set up a model of macrophage polarization, starting from THP-1 monocytes differentiated into macrophages using PMA (Phorbol 12-myristate 13-acetate). Once differentiated (M0 macrophages), they were incubated with IL-4 and IL-13 in order to obtain M2 polarized macrophages or with IFN-gamma and LPS for classical macrophage activation (M1). To mimic the communication between cancer cells and TAMs, M0, M1 or M2 macrophages and HepG2 or A549 cancer cells were co-cultured during respectively 16 (HepG2) or 24 (A549) hours, before etoposide exposure for 24 (HepG2) or 16 (A549) hours. After the incubation, the impact of etoposide on macrophage polarization was studied and cancer cell apoptosis was assessed by western-blot for cleaved caspase-3 and cleaved PARP-1 protein, caspase activity assay and FACS analysis of Annexin V and PI staining.
Results: mRNA and protein expression of M1 and M2 markers confirmed the polarization of THP-1-derived macrophages, which provide a new, easy and well-characterized model of polarized human macrophages. Etoposide-induced cancer cell apoptosis was markedly reduced in the presence of THP-1 M2 macrophages, while apoptosis was increased in cells co-cultured with M1 macrophages. On the other hand, etoposide did not influence M1 or M2 polarization.
Conclusions: These results evidence for the first time a clear protective effect of M2 on the contrary to M1 macrophages on etoposide-induced cancer cell apoptosis.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

