Post-Transcriptional Coordination of the Arabidopsis Iron Deficiency Response is Partially Dependent on the E3 Ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2)
- PMID: 26253232
- PMCID: PMC4597148
- DOI: 10.1074/mcp.M115.048520
Post-Transcriptional Coordination of the Arabidopsis Iron Deficiency Response is Partially Dependent on the E3 Ligases RING DOMAIN LIGASE1 (RGLG1) and RING DOMAIN LIGASE2 (RGLG2)
Abstract
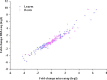
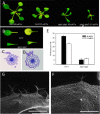
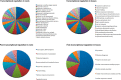
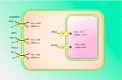
Acclimation to changing environmental conditions is mediated by proteins, the abundance of which is carefully tuned by an elaborate interplay of DNA-templated and post-transcriptional processes. To dissect the mechanisms that control and mediate cellular iron homeostasis, we conducted quantitative high-resolution iTRAQ proteomics and microarray-based transcriptomic profiling of iron-deficient Arabidopsis thaliana plants. A total of 13,706 and 12,124 proteins was identified with a quadrupole-Orbitrap hybrid mass spectrometer in roots and leaves, respectively. This deep proteomic coverage allowed accurate estimates of post-transcriptional regulation in response to iron deficiency. Similarly regulated transcripts were detected in only 13% (roots) and 11% (leaves) of the 886 proteins that differentially accumulated between iron-sufficient and iron-deficient plants, indicating that the majority of the iron-responsive proteins was post-transcriptionally regulated. Mutants harboring defects in the RING DOMAIN LIGASE1 (RGLG1)(1) and RING DOMAIN LIGASE2 (RGLG2) showed a pleiotropic phenotype that resembled iron-deficient plants with reduced trichome density and the formation of branched root hairs. Proteomic and transcriptomic profiling of rglg1 rglg2 double mutants revealed that the functional RGLG protein is required for the regulation of a large set of iron-responsive proteins including the coordinated expression of ribosomal proteins. This integrative analysis provides a detailed catalog of post-transcriptionally regulated proteins and allows the concept of a chiefly transcriptionally regulated iron deficiency response to be revisited. Protein data are available via ProteomeXchange with identifier PXD002126.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

