Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis
- PMID: 26253407
- PMCID: PMC4582183
- DOI: 10.1242/dev.124586
Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis
Abstract
In animals, specification of the primordial germ cells (PGCs), the stem cells of the germ line, is required to transmit genetic information from one generation to the next. Bucky ball (Buc) is essential for germ plasm (GP) assembly in oocytes, and its overexpression results in excess PGCs in zebrafish embryos. However, the mechanistic basis for the excess PGCs in response to Buc overexpression, and whether endogenous Buc functions during embryogenesis, are unknown. Here, we show that endogenous Buc, like GP and overexpressed Buc-GFP, accumulates at embryonic cleavage furrows. Furthermore, we show that the maternally expressed zebrafish Kinesin-1 Kif5Ba is a binding partner of Buc and that maternal kif5Ba (Mkif5Ba) plays an essential role in germline specification in vivo. Specifically, Mkif5Ba is required to recruit GP to cleavage furrows and thereby specifies PGCs. Moreover, Mkif5Ba is required to enrich Buc at cleavage furrows and for the ability of Buc to promote excess PGCs, providing mechanistic insight into how Buc functions to assemble embryonic GP. In addition, we show that Mkif5Ba is also essential for dorsoventral (DV) patterning. Specifically, Mkif5Ba promotes formation of the parallel vegetal microtubule array required to asymmetrically position dorsal determinants (DDs) towards the prospective dorsal side. Interestingly, whereas Syntabulin and wnt8a translocation depend on kif5Ba, grip2a translocation does not, providing evidence for two distinct mechanisms by which DDs might be asymmetrically distributed. These studies identify essential roles for maternal Kif5Ba in PGC specification and DV patterning, and provide mechanistic insight into Buc functions during early embryogenesis.
Keywords: Bucky ball; Dorsoventral patterning; Germ cell; Germ plasm; Kinesin-1; Maternal.
© 2015. Published by The Company of Biologists Ltd.
Figures
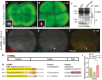

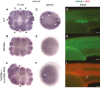
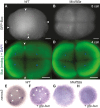
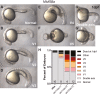


Similar articles
-
A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish.Gene Expr Patterns. 2015 May-Jul;18(1-2):44-52. doi: 10.1016/j.gep.2015.05.003. Epub 2015 Jul 2. Gene Expr Patterns. 2015. PMID: 26143227
-
Bucky ball organizes germ plasm assembly in zebrafish.Curr Biol. 2009 Mar 10;19(5):414-22. doi: 10.1016/j.cub.2009.01.038. Epub 2009 Feb 26. Curr Biol. 2009. PMID: 19249209
-
Multiplicity of Buc copies in Atlantic salmon contrasts with loss of the germ cell determinant in primates, rodents and axolotl.BMC Evol Biol. 2016 Oct 26;16(1):232. doi: 10.1186/s12862-016-0809-7. BMC Evol Biol. 2016. PMID: 27784263 Free PMC article.
-
The maternal coordinate system: Molecular-genetics of embryonic axis formation and patterning in the zebrafish.Curr Top Dev Biol. 2020;140:341-389. doi: 10.1016/bs.ctdb.2020.05.002. Epub 2020 Jun 16. Curr Top Dev Biol. 2020. PMID: 32591080 Review.
-
Localization in Oogenesis of Maternal Regulators of Embryonic Development.Adv Exp Med Biol. 2017;953:173-207. doi: 10.1007/978-3-319-46095-6_5. Adv Exp Med Biol. 2017. PMID: 27975273 Review.
Cited by
-
Functional equivalence of germ plasm organizers.PLoS Genet. 2018 Nov 6;14(11):e1007696. doi: 10.1371/journal.pgen.1007696. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30399145 Free PMC article.
-
Modulation of F-actin dynamics by maternal Mid1ip1L controls germ plasm aggregation and furrow recruitment in the zebrafish embryo.Development. 2018 May 17;145(10):dev156596. doi: 10.1242/dev.156596. Development. 2018. PMID: 29724756 Free PMC article.
-
Kinesin-1 promotes chondrocyte maintenance during skeletal morphogenesis.PLoS Genet. 2017 Jul 17;13(7):e1006918. doi: 10.1371/journal.pgen.1006918. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28715414 Free PMC article.
-
Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish.Commun Biol. 2020 Mar 3;3(1):94. doi: 10.1038/s42003-020-0827-2. Commun Biol. 2020. PMID: 32127635 Free PMC article.
-
Vertebrate Axial Patterning: From Egg to Asymmetry.Adv Exp Med Biol. 2017;953:209-306. doi: 10.1007/978-3-319-46095-6_6. Adv Exp Med Biol. 2017. PMID: 27975274 Free PMC article. Review.
References
-
- Becker K. A. and Hart N. H. (1996). The cortical actin cytoskeleton of unactivated zebrafish eggs: spatial organization and distribution of filamentous actin, nonfilamentous actin, and myosin-II. Mol. Reprod. Dev. 43, 536-547. 10.1002/(SICI)1098-2795(199604)43:4<536::AID-MRD17>3.0.CO;2-X - DOI - PubMed
-
- Becker K. A. and Hart N. H. (1999). Reorganization of filamentous actin and myosin-II in zebrafish eggs correlates temporally and spatially with cortical granule exocytosis. J. Cell Sci. 112, 97-110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

