Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis
- PMID: 26253537
- PMCID: PMC4536313
- DOI: 10.1101/gad.265876.115
Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis
Abstract
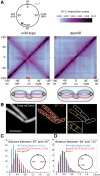
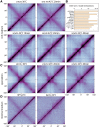
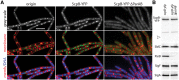
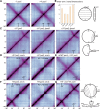
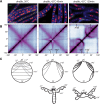
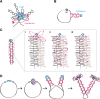
SMC condensin complexes play a central role in compacting and resolving replicated chromosomes in virtually all organisms, yet how they accomplish this remains elusive. In Bacillus subtilis, condensin is loaded at centromeric parS sites, where it encircles DNA and individualizes newly replicated origins. Using chromosome conformation capture and cytological assays, we show that condensin recruitment to origin-proximal parS sites is required for the juxtaposition of the two chromosome arms. Recruitment to ectopic parS sites promotes alignment of large tracks of DNA flanking these sites. Importantly, insertion of parS sites on opposing arms indicates that these "zip-up" interactions only occur between adjacent DNA segments. Collectively, our data suggest that condensin resolves replicated origins by promoting the juxtaposition of DNA flanking parS sites, drawing sister origins in on themselves and away from each other. These results are consistent with a model in which condensin encircles the DNA flanking its loading site and then slides down, tethering the two arms together. Lengthwise condensation via loop extrusion could provide a generalizable mechanism by which condensin complexes act dynamically to individualize origins in B. subtilis and, when loaded along eukaryotic chromosomes, resolve them during mitosis.
Keywords: DNA segregation; Hi-C; ParB; SMC; lengthwise condensation; mitosis; parS.
© 2015 Wang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536. - PubMed
-
- Boonstra M, de Jong IG, Scholefield G, Murray H, Kuipers OP, Veening JW. 2013. Spo0A regulates chromosome copy number during sporulation by directly binding to the origin of replication in Bacillus subtilis. Mol Microbiol 87: 925–938. - PubMed
-
- Breier AM, Grossman AD. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol Microbiol 64: 703–718. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
