The Ribosome-Sec61 Translocon Complex Forms a Cytosolically Restricted Environment for Early Polytopic Membrane Protein Folding
- PMID: 26254469
- PMCID: PMC4661407
- DOI: 10.1074/jbc.M115.672261
The Ribosome-Sec61 Translocon Complex Forms a Cytosolically Restricted Environment for Early Polytopic Membrane Protein Folding
Abstract
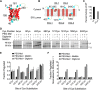
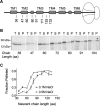
Transmembrane topology of polytopic membrane proteins (PMPs) is established in the endoplasmic reticulum (ER) by the ribosome Sec61-translocon complex (RTC) through iterative cycles of translocation initiation and termination. It remains unknown, however, whether tertiary folding of transmembrane domains begins after the nascent polypeptide integrates into the lipid bilayer or within a proteinaceous environment proximal to translocon components. To address this question, we used cysteine scanning mutagenesis to monitor aqueous accessibility of stalled translation intermediates to determine when, during biogenesis, hydrophilic peptide loops of the aquaporin-4 (AQP4) water channel are delivered to cytosolic and lumenal compartments. Results showed that following ribosome docking on the ER membrane, the nascent polypeptide was shielded from the cytosol as it emerged from the ribosome exit tunnel. Extracellular loops followed a well defined path through the ribosome, the ribosome translocon junction, the Sec61-translocon pore, and into the ER lumen coincident with chain elongation. In contrast, intracellular loops (ICLs) and C-terminalresidues exited the ribosome into a cytosolically shielded environment and remained inaccessible to both cytosolic and lumenal compartments until translation was terminated. Shielding of ICL1 and ICL2, but not the C terminus, became resistant to maneuvers that disrupt electrostatic ribosome interactions. Thus, the early folding landscape of polytopic proteins is shaped by a spatially restricted environment localized within the assembled ribosome translocon complex.
Keywords: aquaporin; endoplasmic reticulum (ER); membrane protein; polytopic membrane protein; protein folding; protein translocation; ribosome function; ribosome translocon complex; sec61 translocon.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Alder N. N., and Johnson A. E. (2004) Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 279, 22787–22790 - PubMed
-
- Hegde R. S., and Lingappa V. R. (1997) Membrane protein biogenesis: regulated complexity at the endoplasmic reticulum. Cell 91, 575–582 - PubMed
-
- White S. H., and von Heijne G. (2004) The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 14, 397–404 - PubMed
-
- Walter P., and Lingappa V. R. (1986) Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 2, 499–516 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

