Deciphering voltage-gated Na(+) and Ca(2+) channels by studying prokaryotic ancestors
- PMID: 26254514
- PMCID: PMC4553089
- DOI: 10.1016/j.tibs.2015.07.002
Deciphering voltage-gated Na(+) and Ca(2+) channels by studying prokaryotic ancestors
Abstract
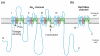
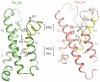
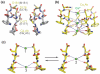
Voltage-gated sodium channels (NaVs) and calcium channels (CaVs) are involved in electrical signaling, contraction, secretion, synaptic transmission, and other physiological processes activated in response to depolarization. Despite their physiological importance, the structures of these closely related proteins have remained elusive because of their size and complexity. Bacterial NaVs have structures analogous to a single domain of eukaryotic NaVs and CaVs and are their likely evolutionary ancestor. Here we review recent work that has led to new understanding of NaVs and CaVs through high-resolution structural studies of their prokaryotic ancestors. New insights into their voltage-dependent activation and inactivation, ion conductance, and ion selectivity provide realistic structural models for the function of these complex membrane proteins at the atomic level.
Keywords: Na(V)Ab; NaChBac; selectivity filter; slow inactivation; voltage sensor; voltage-gated calcium channel; voltage-gated sodium channel.
Published by Elsevier Ltd.
Figures



References
-
- Hille B. Ionic Channels of Excitable Membranes. 3rd Ed Sinauer Associates Inc.; Sunderland, MA: 2001.
-
- Tsien RW. Calcium channels in excitable cell membranes. Annu Rev Physiol. 1983;45:341–358. - PubMed
-
- Catterall WA. The molecular basis of neuronal excitability. Science. 1984;223:653–661. - PubMed
-
- Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

