Systems Analysis of Protein Fatty Acylation in Herpes Simplex Virus-Infected Cells Using Chemical Proteomics
- PMID: 26256475
- PMCID: PMC4543063
- DOI: 10.1016/j.chembiol.2015.06.024
Systems Analysis of Protein Fatty Acylation in Herpes Simplex Virus-Infected Cells Using Chemical Proteomics
Abstract
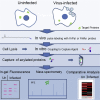
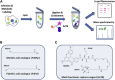
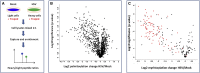
Protein fatty acylation regulates diverse aspects of cellular function and organization and plays a key role in host immune responses to infection. Acylation also modulates the function and localization of virus-encoded proteins. Here, we employ chemical proteomics tools, bio-orthogonal probes, and capture reagents to study myristoylation and palmitoylation during infection with herpes simplex virus (HSV). Using in-gel fluorescence imaging and quantitative mass spectrometry, we demonstrate a generalized reduction in myristoylation of host proteins, whereas palmitoylation of host proteins, including regulators of interferon and tetraspanin family proteins, was selectively repressed. Furthermore, we found that a significant fraction of the viral proteome undergoes palmitoylation; we identified a number of virus membrane glycoproteins, structural proteins, and kinases. Taken together, our results provide broad oversight of protein acylation during HSV infection, a roadmap for similar analysis in other systems, and a resource with which to pursue specific analysis of systems and functions.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



Similar articles
-
Bioorthogonal mimetics of palmitoyl-CoA and myristoyl-CoA and their subsequent isolation by click chemistry and characterization by mass spectrometry reveal novel acylated host-proteins modified by HIV-1 infection.Proteomics. 2015 Jun;15(12):2066-77. doi: 10.1002/pmic.201500063. Epub 2015 May 26. Proteomics. 2015. PMID: 25914232
-
Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection.Mol Cell Proteomics. 2017 Apr;16(4 suppl 1):S92-S107. doi: 10.1074/mcp.M116.065987. Epub 2017 Feb 8. Mol Cell Proteomics. 2017. PMID: 28179408 Free PMC article.
-
Chemoproteomics reveals Toll-like receptor fatty acylation.BMC Biol. 2014 Nov 5;12:91. doi: 10.1186/s12915-014-0091-3. BMC Biol. 2014. PMID: 25371237 Free PMC article.
-
Proteome-wide analysis of protein lipidation using chemical probes: in-gel fluorescence visualization, identification and quantification of N-myristoylation, N- and S-acylation, O-cholesterylation, S-farnesylation and S-geranylgeranylation.Nat Protoc. 2021 Nov;16(11):5083-5122. doi: 10.1038/s41596-021-00601-6. Epub 2021 Oct 27. Nat Protoc. 2021. PMID: 34707257 Review.
-
A Decade of Click Chemistry in Protein Palmitoylation: Impact on Discovery and New Biology.Cell Chem Biol. 2018 Mar 15;25(3):236-246. doi: 10.1016/j.chembiol.2017.12.002. Epub 2017 Dec 28. Cell Chem Biol. 2018. PMID: 29290622 Review.
Cited by
-
Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies.Chem Rev. 2018 Feb 14;118(3):919-988. doi: 10.1021/acs.chemrev.6b00750. Epub 2018 Jan 2. Chem Rev. 2018. PMID: 29292991 Free PMC article.
-
Tracing the STING exocytosis pathway during herpes viruses infection.mBio. 2024 Apr 10;15(4):e0037324. doi: 10.1128/mbio.00373-24. Epub 2024 Mar 12. mBio. 2024. PMID: 38470056 Free PMC article.
-
The function and mechanism of protein acylation in the regulation of viral infection.Virulence. 2025 Dec;16(1):2530171. doi: 10.1080/21505594.2025.2530171. Epub 2025 Jul 17. Virulence. 2025. PMID: 40673681 Free PMC article. Review.
-
Acid Sphingomyelinase regulates the localization and trafficking of palmitoylated proteins.Biol Open. 2019 Oct 15;8(10):bio040311. doi: 10.1242/bio.040311. Biol Open. 2019. PMID: 31142470 Free PMC article.
-
Roles of palmitoylation in axon growth, degeneration and regeneration.J Neurosci Res. 2017 Aug;95(8):1528-1539. doi: 10.1002/jnr.24003. Epub 2017 Feb 2. J Neurosci Res. 2017. PMID: 28150429 Free PMC article. Review.
References
-
- Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta. 2011;1808:2981–2994. - PubMed
-
- Berry A.F.H., Heal W.P., Tarafder A.K., Tolmachova T., Baron R.A., Seabra M.C., Tate E.W. Rapid multilabel detection of geranylgeranylated proteins by using bioorthogonal ligation chemistry. Chembiochem. 2010;11:771–773. - PubMed
-
- Bijlmakers M.J. Protein acylation and localization in T cell signaling (Review) Mol. Membr. Biol. 2009;26:93–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

