High-Throughput Multiplexed Peptide-Centric Profiling Illustrates Both Substrate Cleavage Redundancy and Specificity in the MMP Family
- PMID: 26256476
- PMCID: PMC4546507
- DOI: 10.1016/j.chembiol.2015.07.008
High-Throughput Multiplexed Peptide-Centric Profiling Illustrates Both Substrate Cleavage Redundancy and Specificity in the MMP Family
Abstract
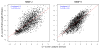
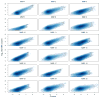
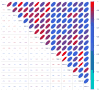
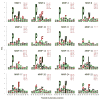
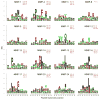
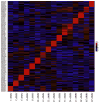
Matrix metalloproteinases (MMPs) play incompletely understood roles in health and disease. Knowing the MMP cleavage preferences is essential for a better understanding of the MMP functions and design of selective inhibitors. To elucidate the cleavage preferences of MMPs, we employed a high-throughput multiplexed peptide-centric profiling technology involving the cleavage of 18,583 peptides by 18 proteinases from the main sub-groups of the MMP family. Our results enabled comparison of the MMP substrates on a global scale, leading to the most efficient and selective substrates. The data validated the accuracy of our cleavage prediction software. This software allows us and others to locate, with nearly 100% accuracy, the MMP cleavage sites in the peptide sequences. In addition to increasing our understanding of both the selectivity and the redundancy of the MMP family, our study generated a roadmap for the subsequent MMP structural-functional studies and efficient substrate and inhibitor design.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
High resolution analysis of snake venom metalloproteinase (SVMP) peptide bond cleavage specificity using proteome based peptide libraries and mass spectrometry.J Proteomics. 2011 Apr 1;74(4):401-10. doi: 10.1016/j.jprot.2010.12.002. Epub 2010 Dec 13. J Proteomics. 2011. PMID: 21156218
-
Matrix metalloproteinases - From the cleavage data to the prediction tools and beyond.Biochim Biophys Acta Mol Cell Res. 2017 Nov;1864(11 Pt A):1952-1963. doi: 10.1016/j.bbamcr.2017.03.010. Epub 2017 Mar 24. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28347746 Free PMC article. Review.
-
Substrate hydrolysis by matrix metalloproteinase-9.J Biol Chem. 2001 Jun 8;276(23):20572-8. doi: 10.1074/jbc.M100900200. Epub 2001 Mar 14. J Biol Chem. 2001. PMID: 11279151
-
Degradation of tropoelastin by matrix metalloproteinases--cleavage site specificities and release of matrikines.FEBS J. 2010 Apr;277(8):1939-56. doi: 10.1111/j.1742-4658.2010.07616.x. Epub 2010 Mar 22. FEBS J. 2010. PMID: 20345904
-
Crystal structures of MMPs in complex with physiological and pharmacological inhibitors.Biochimie. 2005 Mar-Apr;87(3-4):249-63. doi: 10.1016/j.biochi.2004.11.019. Biochimie. 2005. PMID: 15781312 Review.
Cited by
-
PepSeq: a fully in vitro platform for highly multiplexed serology using customizable DNA-barcoded peptide libraries.Nat Protoc. 2023 Feb;18(2):396-423. doi: 10.1038/s41596-022-00766-8. Epub 2022 Nov 16. Nat Protoc. 2023. PMID: 36385198 Free PMC article. Review.
-
Kallikrein-Related Peptidase 14 Activates Zymogens of Membrane Type Matrix Metalloproteinases (MT-MMPs)-A CleavEx Based Analysis.Int J Mol Sci. 2020 Jun 19;21(12):4383. doi: 10.3390/ijms21124383. Int J Mol Sci. 2020. PMID: 32575583 Free PMC article.
-
Both Drosophila matrix metalloproteinases have released and membrane-tethered forms but have different substrates.Sci Rep. 2017 Mar 16;7:44560. doi: 10.1038/srep44560. Sci Rep. 2017. PMID: 28300207 Free PMC article.
-
Matrix Metalloproteinase Triple-Helical Peptide Inhibitors: Potential Cross-Reactivity with Caspase-11.Molecules. 2019 Nov 28;24(23):4355. doi: 10.3390/molecules24234355. Molecules. 2019. PMID: 31795279 Free PMC article.
-
A cell surface display fluorescent biosensor for measuring MMP14 activity in real-time.Sci Rep. 2018 Apr 12;8(1):5916. doi: 10.1038/s41598-018-24080-0. Sci Rep. 2018. PMID: 29651043 Free PMC article.
References
-
- Bauvois B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim Biophys Acta. 2012;1825:29–36. - PubMed
-
- Belkin AM, Akimov SS, Zaritskaya LS, Ratnikov BI, Deryugina EI, Strongin AY. Matrix-dependent proteolysis of surface transglutaminase by membrane- type metalloproteinase regulates cancer cell adhesion and locomotion. J Biol Chem. 2001;276:18415–18422. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

