Hierarchical Clustering With Prototypes via Minimax Linkage
- PMID: 26257451
- PMCID: PMC4527350
- DOI: 10.1198/jasa.2011.tm10183
Hierarchical Clustering With Prototypes via Minimax Linkage
Abstract
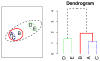
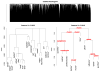
Agglomerative hierarchical clustering is a popular class of methods for understanding the structure of a dataset. The nature of the clustering depends on the choice of linkage-that is, on how one measures the distance between clusters. In this article we investigate minimax linkage, a recently introduced but little-studied linkage. Minimax linkage is unique in naturally associating a prototype chosen from the original dataset with every interior node of the dendrogram. These prototypes can be used to greatly enhance the interpretability of a hierarchical clustering. Furthermore, we prove that minimax linkage has a number of desirable theoretical properties; for example, minimax-linkage dendrograms cannot have inversions (unlike centroid linkage) and is robust against certain perturbations of a dataset. We provide an efficient implementation and illustrate minimax linkage's strengths as a data analysis and visualization tool on a study of words from encyclopedia articles and on a dataset of images of human faces.
Keywords: Agglomerative; Dendrogram; Unsupervised learning.
Figures








References
-
- Ao SI, Yip K, Ng M, Cheung D, Fong P-Y, Melhado I, Sham PC. Clustag: Hierarchical Clustering and Graph Methods for Selecting Tag Snps. Bioinformatics. 2005;21(8):1735–1736. - PubMed
-
- Basalto N, Bellotti R, De Carlo F, Facchi P, Pantaleo E, Pascazio S. Hausdorff Clustering. Physical Review E. 2008;78(4):046112. - PubMed
-
- Bellman RE. Adaptive Control Processes. Princeton, NJ: Princeton University Press; 1961.
-
- Chipman H, Tibshirani R. Hybrid Hierarchical Clustering With Applications to Microarray Data. Biostatistics. 2005;7:286–301. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
