Reconstruction of the temporal signaling network in Salmonella-infected human cells
- PMID: 26257716
- PMCID: PMC4507143
- DOI: 10.3389/fmicb.2015.00730
Reconstruction of the temporal signaling network in Salmonella-infected human cells
Abstract
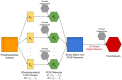
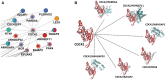
Salmonella enterica is a bacterial pathogen that usually infects its host through food sources. Translocation of the pathogen proteins into the host cells leads to changes in the signaling mechanism either by activating or inhibiting the host proteins. Given that the bacterial infection modifies the response network of the host, a more coherent view of the underlying biological processes and the signaling networks can be obtained by using a network modeling approach based on the reverse engineering principles. In this work, we have used a published temporal phosphoproteomic dataset of Salmonella-infected human cells and reconstructed the temporal signaling network of the human host by integrating the interactome and the phosphoproteomic dataset. We have combined two well-established network modeling frameworks, the Prize-collecting Steiner Forest (PCSF) approach and the Integer Linear Programming (ILP) based edge inference approach. The resulting network conserves the information on temporality, direction of interactions, while revealing hidden entities in the signaling, such as the SNARE binding, mTOR signaling, immune response, cytoskeleton organization, and apoptosis pathways. Targets of the Salmonella effectors in the host cells such as CDC42, RHOA, 14-3-3δ, Syntaxin family, Oxysterol-binding proteins were included in the reconstructed signaling network although they were not present in the initial phosphoproteomic data. We believe that integrated approaches, such as the one presented here, have a high potential for the identification of clinical targets in infectious diseases, especially in the Salmonella infections.
Keywords: Salmonella infection; network reconstruction; pathway analysis; phosphoproteomic; temporal data integration.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

