Remote homology and the functions of metagenomic dark matter
- PMID: 26257768
- PMCID: PMC4508852
- DOI: 10.3389/fgene.2015.00234
Remote homology and the functions of metagenomic dark matter
Abstract
Predicted open reading frames (ORFs) that lack detectable homology to known proteins are termed ORFans. Despite their prevalence in metagenomes, the extent to which ORFans encode real proteins, the degree to which they can be annotated, and their functional contributions, remain unclear. To gain insights into these questions, we applied sensitive remote-homology detection methods to functionally analyze ORFans from soil, marine, and human gut metagenome collections. ORFans were identified, clustered into sequence families, and annotated through profile-profile comparison to proteins of known structure. We found that a considerable number of metagenomic ORFans (73,896 of 484,121, 15.3%) exhibit significant remote homology to structurally characterized proteins, providing a means for ORFan functional profiling. The extent of detected remote homology far exceeds that obtained for artificial protein families (1.4%). As expected for real genes, the predicted functions of ORFans are significantly similar to the functions of their gene neighbors (p < 0.001). Compared to the functional profiles predicted through standard homology searches, ORFans show biologically intriguing differences. Many ORFan-enriched functions are virus-related and tend to reflect biological processes associated with extreme sequence diversity. Each environment also possesses a large number of unique ORFan families and functions, including some known to play important community roles such as gut microbial polysaccharide digestion. Lastly, ORFans are a valuable resource for finding novel enzymes of interest, as we demonstrate through the identification of hundreds of novel ORFan metalloproteases that all possess a signature catalytic motif despite a general lack of similarity to known proteins. Our ORFan functional predictions are a valuable resource for discovering novel protein families and exploring the boundaries of protein sequence space. All remote homology predictions are available at http://doxey.uwaterloo.ca/ORFans.
Keywords: ORFan; comparative metagenomics; functional annotation; metagenome; metaproteome; orphan; profile-profile comparison; remote homology.
Figures
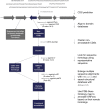
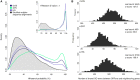
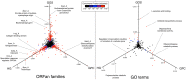

References
-
- Böttger A., Doxey A. C., Hess M. W., Pfaller K., Salvenmoser W., Deutzmann R., et al. (2012). Horizontal gene transfer contributed to the evolution of extracellular surface structures: the freshwater polyp Hydra is covered by a complex fibrous cuticle containing glycosaminoglycans and proteins of the PPOD and SWT (sweet tooth) families. PLoS ONE 7:e52278 10.1371/journal.pone.0052278 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

