SpeedSeq: ultra-fast personal genome analysis and interpretation
- PMID: 26258291
- PMCID: PMC4589466
- DOI: 10.1038/nmeth.3505
SpeedSeq: ultra-fast personal genome analysis and interpretation
Abstract
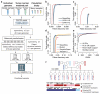
SpeedSeq is an open-source genome analysis platform that accomplishes alignment, variant detection and functional annotation of a 50× human genome in 13 h on a low-cost server and alleviates a bioinformatics bottleneck that typically demands weeks of computation with extensive hands-on expert involvement. SpeedSeq offers performance competitive with or superior to current methods for detecting germline and somatic single-nucleotide variants, structural variants, insertions and deletions, and it includes novel functionality for streamlined interpretation.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

