Prediction of human population responses to toxic compounds by a collaborative competition
- PMID: 26258538
- PMCID: PMC4568441
- DOI: 10.1038/nbt.3299
Prediction of human population responses to toxic compounds by a collaborative competition
Erratum in
-
Erratum: Prediction of human population responses to toxic compounds by a collaborative competition.Nat Biotechnol. 2015 Oct;33(10):1109. doi: 10.1038/nbt1015-1109a. Nat Biotechnol. 2015. PMID: 26448092 Free PMC article. No abstract available.
Abstract
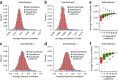
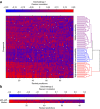
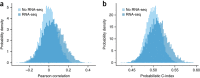
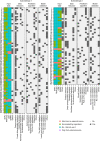
The ability to computationally predict the effects of toxic compounds on humans could help address the deficiencies of current chemical safety testing. Here, we report the results from a community-based DREAM challenge to predict toxicities of environmental compounds with potential adverse health effects for human populations. We measured the cytotoxicity of 156 compounds in 884 lymphoblastoid cell lines for which genotype and transcriptional data are available as part of the Tox21 1000 Genomes Project. The challenge participants developed algorithms to predict interindividual variability of toxic response from genomic profiles and population-level cytotoxicity data from structural attributes of the compounds. 179 submitted predictions were evaluated against an experimental data set to which participants were blinded. Individual cytotoxicity predictions were better than random, with modest correlations (Pearson's r < 0.28), consistent with complex trait genomic prediction. In contrast, predictions of population-level response to different compounds were higher (r < 0.66). The results highlight the possibility of predicting health risks associated with unknown compounds, although risk estimation accuracy remains suboptimal.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

