RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms
- PMID: 26258787
- PMCID: PMC4576190
- DOI: 10.3390/v7082829
RNA-Dependent RNA Polymerases of Picornaviruses: From the Structure to Regulatory Mechanisms
Abstract
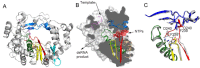
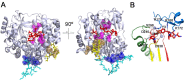
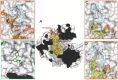
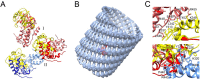
RNA viruses typically encode their own RNA-dependent RNA polymerase (RdRP) to ensure genome replication within the infected cells. RdRP function is critical not only for the virus life cycle but also for its adaptive potential. The combination of low fidelity of replication and the absence of proofreading and excision activities within the RdRPs result in high mutation frequencies that allow these viruses a rapid adaptation to changing environments. In this review, we summarize the current knowledge about structural and functional aspects on RdRP catalytic complexes, focused mainly in the Picornaviridae family. The structural data currently available from these viruses provided high-resolution snapshots for a range of conformational states associated to RNA template-primer binding, rNTP recognition, catalysis and chain translocation. As these enzymes are major targets for the development of antiviral compounds, such structural information is essential for the design of new therapies.
Keywords: RNA-dependent RNA polymerase; picornaviruses; positive-strand RNA viruses; replication fidelity; viral replication.
Figures


References
-
- Wimmer E., Paul A.V. The making of a picornavirus genome. In: Ehrenfeld E., Domingo E., Ross R.P., editors. The Picornavirus. ASM Press; Washington, DC, USA: 2010. pp. 33–55.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

