Expression of the Helicobacter pylori virulence factor vacuolating cytotoxin A (vacA) is influenced by a potential stem-loop structure in the 5' untranslated region of the transcript
- PMID: 26259667
- PMCID: PMC4843948
- DOI: 10.1111/mmi.13160
Expression of the Helicobacter pylori virulence factor vacuolating cytotoxin A (vacA) is influenced by a potential stem-loop structure in the 5' untranslated region of the transcript
Abstract
The vacuolating cytotoxin, VacA, is an important virulence factor secreted by the gastric pathogen Helicobacter pylori. Certain vacA genotypes are strongly associated with disease risk, but the association is not absolute. The factors determining vacA gene expression are not fully understood, and the mechanisms of its regulation are elusive. We have identified a potential mRNA stem-loop forming structure in the 5' untranslated region (UTR) of the vacA transcript. Using site-directed mutagenesis, we found that disruption of the stem-loop structure reduced steady-state mRNA levels between two- and sixfold (P = 0.0005) and decreased mRNA half-life compared with wild type (P = 0.03). This led to a marked reduction in VacA protein levels and overall toxin activity. Additionally, during stressful environmental conditions of acid pH or high environmental salt concentrations, when general transcription of vacA was decreased or increased respectively, the stabilising effects of the stem-loop were even more pronounced. Our results suggest that the stem-loop structure in the vacA 5' UTR is an important determinant of vacA expression through stabilisation of the vacA mRNA transcript and that the stabilising effect is of particular importance during conditions of environmental stress.
© 2015 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures



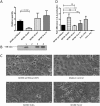
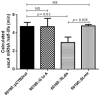
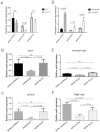

References
-
- Allan, E. , Clayton, C.L. , McLaren, A. , Wallace, D.M. , and Wren, B.W. (2001) Characterization of the low‐pH responses of Helicobacter pylori using genomic DNA arrays. Microbiology 147: 2285–2292. - PubMed
-
- Atherton, J.C. , Cao, P. , Peek, R.M., Jr , Tummuru, M.K. , Blaser, M.J. , and Cover, T.L. (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem 270: 17771–17777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

