Role of complement receptor 1 (CR1; CD35) on epithelial cells: A model for understanding complement-mediated damage in the kidney
- PMID: 26260209
- PMCID: PMC4565762
- DOI: 10.1016/j.molimm.2015.07.016
Role of complement receptor 1 (CR1; CD35) on epithelial cells: A model for understanding complement-mediated damage in the kidney
Abstract
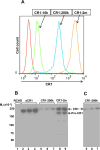
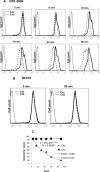
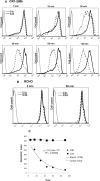
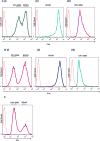
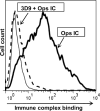
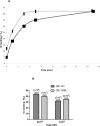
The regulators of complement activation gene cluster encodes a group of proteins that have evolved to control the amplification of complement at the critical step of C3 activation. Complement receptor 1 (CR1) is the most versatile of these inhibitors with both receptor and regulatory functions. While expressed on most peripheral blood cells, the only epithelial site of expression in the kidney is by the podocyte. Its expression by this cell population has aroused considerable speculation as to its biologic function in view of many complement-mediated renal diseases. The goal of this investigation was to assess the role of CR1 on epithelial cells. To this end, we utilized a Chinese hamster ovary cell model system. Among our findings, CR1 reduced C3b deposition by ∼ 80% during classical pathway activation; however, it was an even more potent regulator (>95% reduction in C3b deposition) of the alternative pathway. This inhibition was primarily mediated by decay accelerating activity. The deposited C4b and C3b were progressively cleaved with a t½ of ∼ 30 min to C4d and C3d, respectively, by CR1-dependent cofactor activity. CR1 functioned intrinsically (i.e, worked only on the cell on which it was expressed). Moreover, CR1 efficiently and stably bound but didn't internalize C4b/C3b opsonized immune complexes. Our studies underscore the potential importance of CR1 on an epithelial cell population as both an intrinsic complement regulator and an immune adherence receptor. These results provide a framework for understanding how loss of CR1 expression on podocytes may contribute to complement-mediated damage in the kidney.
Keywords: Cofactor activity; Complement activation; Complement receptor 1; Decay accelerating activity; Epithelial cells; Immune complexes.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures


References
-
- Ahearn JM, Fearon DT. Structure and function of the complement receptors, CR1 (CD35) and CR2 (CD21) Adv Immunol. 1989;46:183–219. - PubMed
-
- Appay MD, Kazatchkine MD, Levi-Strauss M, Hinglais N, Bariety J. Expression of CR1 (CD35) mRNA in podocytes from adult and fetal human kidneys. Kidney Int. 1990;38:289–293. - PubMed
-
- Barilla-LaBarca ML, Liszewski MK, Lambris JD, Hourcade D, Atkinson JP. Role of membrane cofactor protein (CD46) in regulation of C4b and C3b deposited on cells. J Immunol. 2002;168:6298–6304. - PubMed
-
- Brodbeck WG, Mold C, Atkinson JP, Medof ME. Cooperation between decay-accelerating factor and membrane cofactor protein in protecting cells from autologous complement attack. J Immunol. 2000;165:3999–4006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

