Resolving bundled microtubules using anti-tubulin nanobodies
- PMID: 26260773
- PMCID: PMC4918323
- DOI: 10.1038/ncomms8933
Resolving bundled microtubules using anti-tubulin nanobodies
Abstract
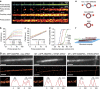
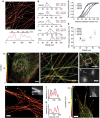
Microtubules are hollow biopolymers of 25-nm diameter and are key constituents of the cytoskeleton. In neurons, microtubules are organized differently between axons and dendrites, but their precise organization in different compartments is not completely understood. Super-resolution microscopy techniques can detect specific structures at an increased resolution, but the narrow spacing between neuronal microtubules poses challenges because most existing labelling strategies increase the effective microtubule diameter by 20-40 nm and will thereby blend neighbouring microtubules into one structure. Here we develop single-chain antibody fragments (nanobodies) against tubulin to achieve super-resolution imaging of microtubules with a decreased apparent diameter. To test the resolving power of these novel probes, we generate microtubule bundles with a known spacing of 50-70 nm and successfully resolve individual microtubules. Individual bundled microtubules can also be resolved in different mammalian cells, including hippocampal neurons, allowing novel insights into fundamental mechanisms of microtubule organization in cell- and neurobiology.
Figures
References
-
- Kapitein L. C. & Hoogenraad C. C. Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol. Cell Neurosci. 46, 9–20 (2011). - PubMed
-
- Chen J., Kanai Y., Cowan N. J. & Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 360, 674–677 (1992). - PubMed
-
- Ries J., Kaplan C., Platonova E., Eghlidi H. & Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012). - PubMed
-
- Royle S. J. Super-duper resolution imaging of mitotic microtubules. Nat. Rev. Mol. Cell Biol. 16, 67 (2015). - PubMed
-
- Nizak C. et al. Recombinant antibodies against subcellular fractions used to track endogenous Golgi protein dynamics in vivo. Traffic 4, 739–753 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

