Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1
- PMID: 26260980
- PMCID: PMC4565128
- DOI: 10.1021/acs.biochem.5b00754
Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1
Abstract
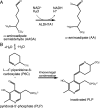
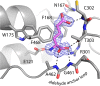
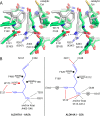
Aldehyde dehydrogenase 7A1 (ALDH7A1) is part of lysine catabolism and catalyzes the NAD(+)-dependent oxidation of α-aminoadipate semialdehyde to α-aminoadipate. Herein, we describe a structural study of human ALDH7A1 focused on substrate recognition. Five crystal structures and small-angle X-ray scattering data are reported, including the first crystal structure of any ALDH7 family member complexed with α-aminoadipate. The product binds with the ε-carboxylate in the oxyanion hole, the aliphatic chain packed into an aromatic box, and the distal end of the product anchored by electrostatic interactions with five conserved residues. This binding mode resembles that of glutamate bound to the proline catabolic enzyme ALDH4A1. Analysis of ALDH7A1 and ALDH4A1 structures suggests key interactions that underlie substrate discrimination. Structures of apo ALDH7A1 reveal dramatic conformational differences from the product complex. Product binding is associated with a 16 Å movement of the C-terminus into the active site, which stabilizes the active conformation of the aldehyde substrate anchor loop. The fact that the C-terminus is part of the active site was hitherto unknown. Interestingly, the C-terminus and aldehyde anchor loop are disordered in a new tetragonal crystal form of the apoenzyme, implying that these parts of the enzyme are highly flexible. Our results suggest that the active site of ALDH7A1 is disassembled when the aldehyde site is vacant, and the C-terminus is a mobile element that forms quaternary structural interactions that aid aldehyde binding. These results are relevant to the c.1512delG genetic deletion associated with pyridoxine-dependent epilepsy, which alters the C-terminus of ALDH7A1.
Figures




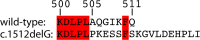
Similar articles
-
Importance of the C-Terminus of Aldehyde Dehydrogenase 7A1 for Oligomerization and Catalytic Activity.Biochemistry. 2017 Nov 7;56(44):5910-5919. doi: 10.1021/acs.biochem.7b00803. Biochemistry. 2017. PMID: 29045138 Free PMC article.
-
Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.FEBS J. 2020 Jan;287(1):173-189. doi: 10.1111/febs.14997. Epub 2019 Jul 25. FEBS J. 2020. PMID: 31302938 Free PMC article.
-
Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function.J Biol Chem. 2016 Nov 11;291(46):24065-24075. doi: 10.1074/jbc.M116.756965. Epub 2016 Sep 27. J Biol Chem. 2016. PMID: 27679491 Free PMC article.
-
Impact of missense mutations in the ALDH7A1 gene on enzyme structure and catalytic function.Biochimie. 2021 Apr;183:49-54. doi: 10.1016/j.biochi.2020.09.016. Epub 2020 Sep 19. Biochimie. 2021. PMID: 32956737 Free PMC article. Review.
-
Aldehyde dehydrogenase and acetaldehyde metabolism.Alcohol Alcohol Suppl. 1994;2:141-5. Alcohol Alcohol Suppl. 1994. PMID: 8974328 Review.
Cited by
-
Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid.Sci Rep. 2016 Oct 19;6:35710. doi: 10.1038/srep35710. Sci Rep. 2016. PMID: 27759097 Free PMC article.
-
The plant pathogen enzyme AldC is a long-chain aliphatic aldehyde dehydrogenase.J Biol Chem. 2020 Oct 2;295(40):13914-13926. doi: 10.1074/jbc.RA120.014747. Epub 2020 Aug 12. J Biol Chem. 2020. PMID: 32796031 Free PMC article.
-
Structural analysis of prolines and hydroxyprolines binding to the l-glutamate-γ-semialdehyde dehydrogenase active site of bifunctional proline utilization A.Arch Biochem Biophys. 2021 Feb 15;698:108727. doi: 10.1016/j.abb.2020.108727. Epub 2020 Dec 18. Arch Biochem Biophys. 2021. PMID: 33333077 Free PMC article.
-
N-carboxyacyl and N-α-aminoacyl derivatives of aminoaldehydes as shared substrates of plant aldehyde dehydrogenases 10 and 7.Amino Acids. 2024 Aug 29;56(1):52. doi: 10.1007/s00726-024-03415-4. Amino Acids. 2024. PMID: 39207552 Free PMC article.
-
Modeling and Structure Determination of Homo-Oligomeric Proteins: An Overview of Challenges and Current Approaches.Int J Mol Sci. 2021 Aug 23;22(16):9081. doi: 10.3390/ijms22169081. Int J Mol Sci. 2021. PMID: 34445785 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

