TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways
- PMID: 26261209
- PMCID: PMC4605304
- DOI: 10.1093/nar/gkv814
TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways
Abstract
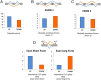
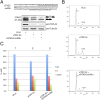
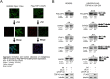
In recent times, high-throughput screening analyses have broadly defined the RNA cellular targets of TDP-43, a nuclear factor involved in neurodegeneration. A common outcome of all these studies is that changing the expression levels of this protein can alter the expression of several hundred RNAs within cells. What still remains to be clarified is which changes represent direct cellular targets of TDP-43 or just secondary variations due to the general role played by this protein in RNA metabolism. Using an HTS-based splicing junction analysis we identified at least six bona fide splicing events that are consistent with being controlled by TDP-43. Validation of the data, both in neuronal and non-neuronal cell lines demonstrated that TDP-43 substantially alters the levels of isoform expression in four genes potentially important for neuropathology: MADD/IG20, STAG2, FNIP1 and BRD8. For MADD/IG20 and STAG2, these changes could also be confirmed at the protein level. These alterations were also observed in a cellular model that successfully mimics TDP-43 loss of function effects following its aggregation. Most importantly, our study demonstrates that cell cycle alterations induced by TDP-43 knockdown can be recovered by restoring the STAG2, an important component of the cohesin complex, normal splicing profile.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
-
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. - PubMed
-
- Buratti E., Baralle F.E. TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 2012;37:237–247. - PubMed
-
- Romano M., Buratti E. Targeting RNA binding proteins involved in neurodegeneration. J. Biomol. Screen. 2013;18:967–983. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

