UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP
- PMID: 26265139
- PMCID: PMC7780642
- DOI: 10.1016/j.bcp.2015.08.084
UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP
Abstract
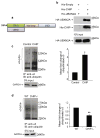
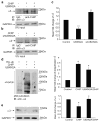
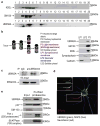
Neuronal nicotinic acetylcholine receptors (nAChRs) containing the α3 subunit are known for their prominent role in normal ganglionic transmission while their involvement in the mechanisms underlying nicotine addiction and smoking-related disease has been emerging only in recent years. The amount of information available on the maturation and trafficking of α3-containing nAChRs is limited. We previously showed that UBXN2A is a p97 adaptor protein that facilitates the maturation and trafficking of α3-containing nAChRs. Further investigation of the mechanisms of UBXN2A actions revealed that the protein interacts with CHIP (carboxyl terminus of Hsc70 interacting protein), whose ubiquitin E3 ligase activity regulates the degradation of several disease-related proteins. We show that CHIP displays E3 ligase activity toward the α3 nAChR subunit and contributes to its ubiquitination and subsequent degradation. UBXN2A interferes with CHIP-mediated ubiquitination of α3 and protects the nicotinic receptor subunit from endoplasmic reticulum associated degradation (ERAD). UBXN2A also cross-talks with VCP/p97 and HSC70/HSP70 proteins in a complex where α3 is likely to be targeted by CHIP. Overall,we identify CHIP as an E3 ligase for α3 and UBXN2A as a protein that may efficiently regulate the stability of CHIP's client substrates.
Keywords: CHIP; Proteasomal degradation; UBX domain; UBXN2A; p97; α3 Nicotinic subunit.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
UBXN2A enhances CHIP-mediated proteasomal degradation of oncoprotein mortalin-2 in cancer cells.Mol Oncol. 2018 Oct;12(10):1753-1777. doi: 10.1002/1878-0261.12372. Epub 2018 Sep 3. Mol Oncol. 2018. PMID: 30107089 Free PMC article.
-
UBXD4, a UBX-containing protein, regulates the cell surface number and stability of alpha3-containing nicotinic acetylcholine receptors.J Neurosci. 2009 May 27;29(21):6883-96. doi: 10.1523/JNEUROSCI.4723-08.2009. J Neurosci. 2009. PMID: 19474315 Free PMC article.
-
The ubiquitin-proteasome system regulates the stability of neuronal nicotinic acetylcholine receptors.J Mol Neurosci. 2010 Jan;40(1-2):177-84. doi: 10.1007/s12031-009-9272-x. Epub 2009 Aug 20. J Mol Neurosci. 2010. PMID: 19693707 Free PMC article.
-
CHIP: a link between the chaperone and proteasome systems.Cell Stress Chaperones. 2003 Winter;8(4):303-8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. Cell Stress Chaperones. 2003. PMID: 15115282 Free PMC article. Review.
-
Membrane Protein Quantity Control at the Endoplasmic Reticulum.J Membr Biol. 2017 Aug;250(4):379-392. doi: 10.1007/s00232-016-9931-0. Epub 2016 Oct 14. J Membr Biol. 2017. PMID: 27743014 Free PMC article. Review.
Cited by
-
UBXN2A enhances CHIP-mediated proteasomal degradation of oncoprotein mortalin-2 in cancer cells.Mol Oncol. 2018 Oct;12(10):1753-1777. doi: 10.1002/1878-0261.12372. Epub 2018 Sep 3. Mol Oncol. 2018. PMID: 30107089 Free PMC article.
-
Genomic insights for personalised care in lung cancer and smoking cessation: motivating at-risk individuals toward evidence-based health practices.EBioMedicine. 2024 Dec;110:105441. doi: 10.1016/j.ebiom.2024.105441. Epub 2024 Nov 8. EBioMedicine. 2024. PMID: 39520911 Free PMC article.
-
Chaperone-assisted E3 ligase CHIP: A double agent in cancer.Genes Dis. 2021 Sep 1;9(6):1521-1555. doi: 10.1016/j.gendis.2021.08.003. eCollection 2022 Nov. Genes Dis. 2021. PMID: 36157498 Free PMC article. Review.
-
Exploring the role of ubiquitin regulatory X domain family proteins in cancers: bioinformatics insights, mechanisms, and implications for therapy.J Transl Med. 2024 Feb 15;22(1):157. doi: 10.1186/s12967-024-04890-9. J Transl Med. 2024. PMID: 38365777 Free PMC article. Review.
-
Degradation of the stress-responsive enzyme formate dehydrogenase by the RING-type E3 ligase Keep on Going and the ubiquitin 26S proteasome system.Plant Mol Biol. 2018 Feb;96(3):265-278. doi: 10.1007/s11103-017-0691-8. Epub 2017 Dec 21. Plant Mol Biol. 2018. PMID: 29270890
References
-
- WHO. Fact Sheet # 339. 2014. Tobacco World Health Organization.
-
- De Biasi M. Nicotinic receptors autonomic neurons. In: Squire LR, editor. Encyclopedia of Neuroscience. Academic Press; Oxford: 2009. pp. 1135–1139.
-
- Boulter J, O’Shea-Greenfield A, Duvoisin RM, Connolly JG, Wada E, Jensen A, et al. Alpha 3, alpha 5, and beta 4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J Biol Chem. 1990;265:4472–4482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

