The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy
- PMID: 26266977
- PMCID: PMC5018156
- DOI: 10.1038/nature14893
The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy
Abstract
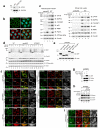
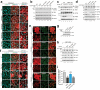
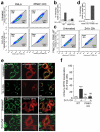
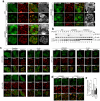
Protein aggregates and damaged organelles are tagged with ubiquitin chains to trigger selective autophagy. To initiate mitophagy, the ubiquitin kinase PINK1 phosphorylates ubiquitin to activate the ubiquitin ligase parkin, which builds ubiquitin chains on mitochondrial outer membrane proteins, where they act to recruit autophagy receptors. Using genome editing to knockout five autophagy receptors in HeLa cells, here we show that two receptors previously linked to xenophagy, NDP52 and optineurin, are the primary receptors for PINK1- and parkin-mediated mitophagy. PINK1 recruits NDP52 and optineurin, but not p62, to mitochondria to activate mitophagy directly, independently of parkin. Once recruited to mitochondria, NDP52 and optineurin recruit the autophagy factors ULK1, DFCP1 and WIPI1 to focal spots proximal to mitochondria, revealing a function for these autophagy receptors upstream of LC3. This supports a new model in which PINK1-generated phospho-ubiquitin serves as the autophagy signal on mitochondria, and parkin then acts to amplify this signal. This work also suggests direct and broader roles for ubiquitin phosphorylation in other autophagy pathways.
Figures









Comment in
-
Cell biology: Tagged tags engage disposal.Nature. 2015 Aug 20;524(7565):294-5. doi: 10.1038/nature15199. Epub 2015 Aug 12. Nature. 2015. PMID: 26266982 No abstract available.
-
Autophagy: Mitophagy receptors unravelled.Nat Rev Mol Cell Biol. 2015 Oct;16(10):580. doi: 10.1038/nrm4058. Epub 2015 Sep 9. Nat Rev Mol Cell Biol. 2015. PMID: 26350072 No abstract available.
-
PINK1, parkin, and autophagy receptors: A new model of mitophagy.Mov Disord. 2016 Nov;31(11):1628-1629. doi: 10.1002/mds.26812. Epub 2016 Oct 6. Mov Disord. 2016. PMID: 27709687 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

